[English] 日本語
 Yorodumi
Yorodumi- PDB-7ut7: C2 symmetric cryoEM structure of Azotobacter vinelandii MoFeP und... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7ut7 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | C2 symmetric cryoEM structure of Azotobacter vinelandii MoFeP under non-turnover conditions | |||||||||||||||
 Components Components | (Nitrogenase molybdenum-iron protein ...) x 2 | |||||||||||||||
 Keywords Keywords | OXIDOREDUCTASE / nitrogenase / MoFeP / nitrogen fixation | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmolybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / nitrogen fixation / iron-sulfur cluster binding / ATP binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 1.91 Å | |||||||||||||||
 Authors Authors | Rutledge, H.L. / Cook, B.D. / Tezcan, F.A. / Herzik, M.A. | |||||||||||||||
| Funding support |  United States, 4items United States, 4items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structures of the nitrogenase complex prepared under catalytic turnover conditions. Authors: Hannah L Rutledge / Brian D Cook / Hoang P M Nguyen / Mark A Herzik / F Akif Tezcan /  Abstract: The enzyme nitrogenase couples adenosine triphosphate (ATP) hydrolysis to the multielectron reduction of atmospheric dinitrogen into ammonia. Despite extensive research, the mechanistic details of ...The enzyme nitrogenase couples adenosine triphosphate (ATP) hydrolysis to the multielectron reduction of atmospheric dinitrogen into ammonia. Despite extensive research, the mechanistic details of ATP-dependent energy transduction and dinitrogen reduction by nitrogenase are not well understood, requiring new strategies to monitor its structural dynamics during catalytic action. Here, we report cryo-electron microscopy structures of the nitrogenase complex prepared under enzymatic turnover conditions. We observe that asymmetry governs all aspects of the nitrogenase mechanism, including ATP hydrolysis, protein-protein interactions, and catalysis. Conformational changes near the catalytic iron-molybdenum cofactor are correlated with the nucleotide-hydrolysis state of the enzyme. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7ut7.cif.gz 7ut7.cif.gz | 404.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7ut7.ent.gz pdb7ut7.ent.gz | 325.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7ut7.json.gz 7ut7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ut/7ut7 https://data.pdbj.org/pub/pdb/validation_reports/ut/7ut7 ftp://data.pdbj.org/pub/pdb/validation_reports/ut/7ut7 ftp://data.pdbj.org/pub/pdb/validation_reports/ut/7ut7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  26757MC  7ut6C  7ut8C  7ut9C  7utaC  8dpnC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Nitrogenase molybdenum-iron protein ... , 2 types, 4 molecules ACBD
| #1: Protein | Mass: 55363.043 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Azotobacter vinelandii DJ (bacteria) / References: UniProt: P07328, nitrogenase Azotobacter vinelandii DJ (bacteria) / References: UniProt: P07328, nitrogenase#2: Protein | Mass: 59535.879 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Azotobacter vinelandii DJ (bacteria) / References: UniProt: C1DGZ8, nitrogenase Azotobacter vinelandii DJ (bacteria) / References: UniProt: C1DGZ8, nitrogenase |
|---|
-Non-polymers , 5 types, 423 molecules 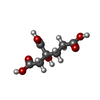
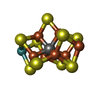



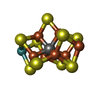



| #3: Chemical | | #4: Chemical | #5: Chemical | #6: Chemical | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Heterotetrameric MoFeP from Azotobacter vinelandii / Type: COMPLEX Details: Wild-type MoFeP was purified from the native organism, Azotobacter vinelandii Entity ID: #1-#2 / Source: NATURAL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.23321 MDa / Experimental value: NO | |||||||||||||||
| Source (natural) | Organism:  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) | |||||||||||||||
| Buffer solution | pH: 8 Details: Solutions were prepared and filtered immediately prior to the experiment. | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Conc.: 1.44 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | |||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 | |||||||||||||||
| Vitrification | Instrument: HOMEMADE PLUNGER / Cryogen name: ETHANE-PROPANE / Humidity: 95 % / Chamber temperature: 277 K / Details: Custom manual plunger. Greater than 95% humidity. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 165000 X / Nominal defocus max: 1500 nm / Nominal defocus min: 500 nm / Cs: 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: ZEMLIN TABLEAU |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Temperature (max): 123 K / Temperature (min): 93 K |
| Image recording | Electron dose: 65 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 2 / Num. of real images: 3773 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.20.1_4487: / Classification: refinement | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING ONLY | ||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 2628629 | ||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 1.91 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 177123 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj




