+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25843 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of GluN1b-2B NMDAR complexed to Fab2 class1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Channel / antibody / SIGNALING PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / sensory organ development / positive regulation of Schwann cell migration / pons maturation / sensitization / regulation of cell communication / EPHB-mediated forward signaling / Assembly and cell surface presentation of NMDA receptors ...cellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / sensory organ development / positive regulation of Schwann cell migration / pons maturation / sensitization / regulation of cell communication / EPHB-mediated forward signaling / Assembly and cell surface presentation of NMDA receptors / response to hydrogen sulfide / auditory behavior / olfactory learning / conditioned taste aversion / dendritic branch / regulation of respiratory gaseous exchange / response to other organism / apical dendrite / regulation of ARF protein signal transduction / fear response / protein localization to postsynaptic membrane / transmitter-gated monoatomic ion channel activity / positive regulation of inhibitory postsynaptic potential / response to methylmercury / suckling behavior / response to manganese ion / response to glycine / interleukin-1 receptor binding / response to carbohydrate / propylene metabolic process / cellular response to dsRNA / cellular response to lipid / response to growth hormone / heterocyclic compound binding / negative regulation of dendritic spine maintenance / positive regulation of glutamate secretion / RAF/MAP kinase cascade / regulation of monoatomic cation transmembrane transport / NMDA glutamate receptor activity / Synaptic adhesion-like molecules / voltage-gated monoatomic cation channel activity / response to glycoside / NMDA selective glutamate receptor complex / glutamate binding / ligand-gated sodium channel activity / neurotransmitter receptor complex / response to morphine / regulation of axonogenesis / neuromuscular process / calcium ion transmembrane import into cytosol / regulation of dendrite morphogenesis / protein heterotetramerization / male mating behavior / regulation of synapse assembly / response to amine / regulation of cAMP/PKA signal transduction / glycine binding / receptor clustering / small molecule binding / startle response / parallel fiber to Purkinje cell synapse / positive regulation of reactive oxygen species biosynthetic process / monoatomic cation transmembrane transport / behavioral response to pain / regulation of MAPK cascade / positive regulation of calcium ion transport into cytosol / cellular response to glycine / extracellularly glutamate-gated ion channel activity / regulation of postsynaptic membrane potential / response to magnesium ion / action potential / associative learning / response to electrical stimulus / excitatory synapse / positive regulation of dendritic spine maintenance / social behavior / monoatomic ion channel complex / monoatomic cation transport / regulation of neuronal synaptic plasticity / glutamate receptor binding / Unblocking of NMDA receptors, glutamate binding and activation / positive regulation of excitatory postsynaptic potential / detection of mechanical stimulus involved in sensory perception of pain / long-term memory / response to mechanical stimulus / neuron development / synaptic cleft / phosphatase binding / prepulse inhibition / positive regulation of synaptic transmission, glutamatergic / behavioral fear response / multicellular organismal response to stress / postsynaptic density, intracellular component / monoatomic cation channel activity / response to fungicide / calcium ion homeostasis / glutamate-gated receptor activity / regulation of long-term synaptic depression / regulation of neuron apoptotic process / cell adhesion molecule binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.92 Å | |||||||||
 Authors Authors | Tajima N / Furukawa H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Development and characterization of functional antibodies targeting NMDA receptors. Authors: Nami Tajima / Noriko Simorowski / Remy A Yovanno / Michael C Regan / Kevin Michalski / Ricardo Gómez / Albert Y Lau / Hiro Furukawa /  Abstract: N-methyl-D-aspartate receptors (NMDARs) are critically involved in basic brain functions and neurodegeneration as well as tumor invasiveness. Targeting specific subtypes of NMDARs with distinct ...N-methyl-D-aspartate receptors (NMDARs) are critically involved in basic brain functions and neurodegeneration as well as tumor invasiveness. Targeting specific subtypes of NMDARs with distinct activities has been considered an effective therapeutic strategy for neurological disorders and diseases. However, complete elimination of off-target effects of small chemical compounds has been challenging and thus, there is a need to explore alternative strategies for targeting NMDAR subtypes. Here we report identification of a functional antibody that specifically targets the GluN1-GluN2B NMDAR subtype and allosterically down-regulates ion channel activity as assessed by electrophysiology. Through biochemical analysis, x-ray crystallography, single-particle electron cryomicroscopy, and molecular dynamics simulations, we show that this inhibitory antibody recognizes the amino terminal domain of the GluN2B subunit and increases the population of the non-active conformational state. The current study demonstrates that antibodies may serve as specific reagents to regulate NMDAR functions for basic research and therapeutic objectives. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25843.map.gz emd_25843.map.gz | 59.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25843-v30.xml emd-25843-v30.xml emd-25843.xml emd-25843.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25843.png emd_25843.png | 58.4 KB | ||
| Filedesc metadata |  emd-25843.cif.gz emd-25843.cif.gz | 7.4 KB | ||
| Others |  emd_25843_additional_1.map.gz emd_25843_additional_1.map.gz emd_25843_half_map_1.map.gz emd_25843_half_map_1.map.gz emd_25843_half_map_2.map.gz emd_25843_half_map_2.map.gz | 59.4 MB 5.1 MB 5.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25843 http://ftp.pdbj.org/pub/emdb/structures/EMD-25843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25843 | HTTPS FTP |
-Validation report
| Summary document |  emd_25843_validation.pdf.gz emd_25843_validation.pdf.gz | 718.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25843_full_validation.pdf.gz emd_25843_full_validation.pdf.gz | 718.3 KB | Display | |
| Data in XML |  emd_25843_validation.xml.gz emd_25843_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_25843_validation.cif.gz emd_25843_validation.cif.gz | 14.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25843 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25843 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25843 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25843 | HTTPS FTP |
-Related structure data
| Related structure data |  7te9MC  7te4C  7te6C  7tebC  7teeC  7teqC  7terC  7tesC  7tetC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25843.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25843.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
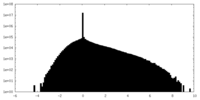
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: sharpened
| File | emd_25843_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
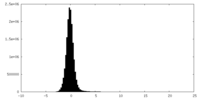
| Density Histograms |
-Half map: #2
| File | emd_25843_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
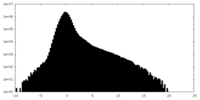
| Density Histograms |
-Half map: #1
| File | emd_25843_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
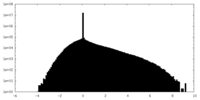
| Density Histograms |
- Sample components
Sample components
-Entire : GluN1b-2B NMDAR - Fab2
| Entire | Name: GluN1b-2B NMDAR - Fab2 |
|---|---|
| Components |
|
-Supramolecule #1: GluN1b-2B NMDAR - Fab2
| Supramolecule | Name: GluN1b-2B NMDAR - Fab2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamate receptor ionotropic, NMDA 1
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 96.944891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTMHLLTFA LLFSCSFARA ASDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL QATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYN W NHIILLVS ...String: MSTMHLLTFA LLFSCSFARA ASDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL QATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYN W NHIILLVS DDHEGRAAQK RLETLLEERE SKSKKRNYEN LDQLSYDNKR GPKAEKVLQF DPGTKNVTAL LMEARELEAR VI ILSASED DAATVYRAAA MLDMTGSGYV WLVGEREISG NALRYAPDGI IGLQLINGKN ESAHISDAVG VVAQAVHELL EKE NITDPP RGCVGNTNIW KTGPLFKRVL MSSKYADGVT GRVEFNEDGD RKFAQYSIMN LQNRKLVQVG IYNGTHVIPN DRKI IWPGG ETEKPRGYQM STRLKIVTIH QEPFVYVKPT MSDGTCKEEF TVNGDPVKKV ICTGPNDTSP GSPRHTVPQC CYGFC IDLL IKLARTMQFT YEVHLVADGK FGTQERVQNS NKKEWNGMMG ELLSGQADMI VAPLTINNER AQYIEFSKPF KYQGLT ILV KKEIPRSTLD SFMQPFQSTL WLLVGLSVHV VAVMLYLLDR FSPFGRFKVN SQSESTDALT LSSAMWFSWG VLLNSGI GE GAPRSFSARI LGMVWAGFAM IIVASYTANL AAFLVLDRPE ERITGINDPR LRNPSDKFIY ATVKQSSVDI YFRRQVEL S TMYRHMEKHN YESAAEAIQA VRDNKLHAFI WDSAVLEFEA SQKCDLVTTG ELFFRSGFGI GMRKDSPWKQ QVSLSILKS HENGFMEDLD KTWVRYQECD SRSNAPATLT CENMAGVFML VAGGIVAGIF LIFIEIAYKS RA UniProtKB: Glutamate receptor ionotropic, NMDA 1 |
-Macromolecule #2: Glutamate receptor ionotropic, NMDA 2B
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 2B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.324406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SPPSIGIAVI LVGTSDEVAI KDAHEKDDFH HLSVVPRVEL VAMNETDPKS IITRICDLMS DRKIQGVVFA DDTDQEAIAQ ILDFISAQT LTPILGIHGG SSMIMADKDE SSMFFQFGPS IEQQASVMLN IMEEYDWYIF SIVTTYFPGY QDFVNKIRST I ENSFVGWE ...String: SPPSIGIAVI LVGTSDEVAI KDAHEKDDFH HLSVVPRVEL VAMNETDPKS IITRICDLMS DRKIQGVVFA DDTDQEAIAQ ILDFISAQT LTPILGIHGG SSMIMADKDE SSMFFQFGPS IEQQASVMLN IMEEYDWYIF SIVTTYFPGY QDFVNKIRST I ENSFVGWE LEEVLLLDMS LDDGDSKIQN QLKKLQSPII LLYCTKEEAT YIFEVANSVG LTGYGYTWIV PSLVAGDTDT VP SEFPTGL ISVSYDEWDY GLPARVRDGI AIITTAASDM LSEHSFIPEP KSSCYNTHEK RIYQSNMLNR YLINVTFEGR DLS FSEDGY QMHPKLVIIL LNKERKWERV GKWKDKSLQM KYYVWPRMCP ETEEQEDDHL SIVTLEEAPF VIVESVDPLS GTCM RNTVP CQKRIISENK TDEEPGYIKK CCKGFCIDIL KKISKSVKFT YDLYLVTNGK HGKKINGTWN GMIGEVVMKR AYMAV GSLT INEERSEVVD FSVPFIETGI SVMVSRSNGT VSPSAFLEPF SACVWVMMFV MLLIVSAVAV FVFEYFSPVG YNRSLA DGR EPGGPSVTIG KAIWLLWGLV FNNSVPVQNP KGTTSKIMVS VWAFFAVIFL ASYTANLAAF MIQEEYVDQV SGLSDKK FQ RPNDFSPPFR FGTVPNGSTE RNIRNNYAEM HAYMGKFNQR GVDDALLSLK TGKLDAFIYD AAVLNYMAGR DEGCKLVT I GSGKVFASTG YGIAIQKDSG WKRQVDLAIL QLFGDGEMEE LEALWLTGIC HNEKNEVMSS QLDIDNMAGV FYMLGAAMA LSLITFISEH LFYWQFRHSF MG UniProtKB: Glutamate receptor ionotropic, NMDA 2B |
-Macromolecule #3: Fab2 heavy chain
| Macromolecule | Name: Fab2 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.914781 KDa |
| Sequence | String: DVKLQESGGG LVQPGGSLKL SCAASGFTFS SYTMSWVRQT PEKRLEWVAY ISNGGGGTYY PDTVKGRFTI SRDNAKNTLY LQMNSLKED TAMYYCARPS RGGSSYWYFD VWGAGTTVTV SSAKTTPPSV YPLAPGSAAQ TNSMVTLGCL VKGYFPEPVT V TWNSGSLS ...String: DVKLQESGGG LVQPGGSLKL SCAASGFTFS SYTMSWVRQT PEKRLEWVAY ISNGGGGTYY PDTVKGRFTI SRDNAKNTLY LQMNSLKED TAMYYCARPS RGGSSYWYFD VWGAGTTVTV SSAKTTPPSV YPLAPGSAAQ TNSMVTLGCL VKGYFPEPVT V TWNSGSLS SGVHTFPAVL QSDLYTLSSS VTVPSSTWPS ETVTCNVAHP ASSTKVDKKI VPRDC |
-Macromolecule #4: Fab2 light chain
| Macromolecule | Name: Fab2 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.572031 KDa |
| Sequence | String: DIQMTQSPSS LSASLGGKVT ITCKASQDIN KYIAWYQHKP GKGPRLLIHY TSSLQPGIPS RFSGSGSGRD YSFSISNLEP EDIATYYCL QYDNLYTFGG GTKLEIKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK ...String: DIQMTQSPSS LSASLGGKVT ITCKASQDIN KYIAWYQHKP GKGPRLLIHY TSSLQPGIPS RFSGSGSGRD YSFSISNLEP EDIATYYCL QYDNLYTFGG GTKLEIKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK DSTYSMSSTL TLTKDEYERH NSYTCEATHK TSTSPIVKSF NRNES |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)