+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of S1PR2-heterotrimeric G13 signaling complex | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | S1P / S1PR2 / GPCR / membrane protein / cryo-EM / SIGNALING PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationD5 dopamine receptor binding / positive regulation of establishment of endothelial barrier / regulation of fibroblast migration / Rho-activating G protein-coupled receptor signaling pathway / sphingosine-1-phosphate receptor activity / Lysosphingolipid and LPA receptors / negative regulation of excitatory postsynaptic potential / filopodium assembly / sphingosine-1-phosphate receptor signaling pathway / G protein-coupled peptide receptor activity ...D5 dopamine receptor binding / positive regulation of establishment of endothelial barrier / regulation of fibroblast migration / Rho-activating G protein-coupled receptor signaling pathway / sphingosine-1-phosphate receptor activity / Lysosphingolipid and LPA receptors / negative regulation of excitatory postsynaptic potential / filopodium assembly / sphingosine-1-phosphate receptor signaling pathway / G protein-coupled peptide receptor activity / regulation of small GTPase mediated signal transduction / positive regulation of peptidyl-threonine phosphorylation / regulation of metabolic process / NRAGE signals death through JNK / branching involved in blood vessel morphogenesis / CDC42 GTPase cycle / regulation of postsynapse assembly / Rho protein signal transduction / RAC1 GTPase cycle / guanyl-nucleotide exchange factor activity / excitatory postsynaptic potential / brush border membrane / G protein-coupled receptor binding / G protein-coupled receptor activity / platelet activation / regulation of blood pressure / integrin binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / melanosome / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / regulation of cell shape / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / presynapse / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / actin cytoskeleton organization / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / in utero embryonic development / Ras protein signal transduction / cell differentiation / Extra-nuclear estrogen signaling / cell population proliferation / postsynapse / G protein-coupled receptor signaling pathway / lysosomal membrane / focal adhesion / GTPase activity / positive regulation of cell population proliferation / synapse / lipid binding / GTP binding / protein-containing complex binding / glutamatergic synapse / signal transduction / extracellular exosome Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.19 Å | ||||||||||||
 Authors Authors | Li X / Chen H | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structure of S1PR2-heterotrimeric G signaling complex. Authors: Hongwen Chen / Kevin Chen / Weijiao Huang / Louis M Staudt / Jason G Cyster / Xiaochun Li /  Abstract: Sphingosine-1-phosphate (S1P) regulates immune cell trafficking, angiogenesis, and vascular function via its five receptors. Inherited mutations in S1P receptor 2 (S1PR2) occur in individuals with ...Sphingosine-1-phosphate (S1P) regulates immune cell trafficking, angiogenesis, and vascular function via its five receptors. Inherited mutations in S1P receptor 2 (S1PR2) occur in individuals with hearing loss, and acquired mutations in S1PR2 and G occur in a malignant lymphoma. Here, we present the cryo-electron microscopy structure of S1P-bound S1PR2 coupled to the heterotrimeric G. Interaction between S1PR2 intracellular loop 2 (ICL2) and transmembrane helix 4 confines ICL2 to engage the α5 helix of G. Transforming growth factor-α shedding assays and cell migration assays support the key roles of the residues in S1PR2-G complex assembly. The structure illuminates the mechanism of receptor disruption by disease-associated mutations. Unexpectedly, we showed that FTY720-P, an agonist of the other four S1PRs, can trigger G activation via S1PR2. S1PR2 variant can increase the activity of G considerably with FTY720-P and S1P, thus revealing a basis for S1PR drug selectivity. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25712.map.gz emd_25712.map.gz | 85.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25712-v30.xml emd-25712-v30.xml emd-25712.xml emd-25712.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25712.png emd_25712.png | 46.8 KB | ||
| Filedesc metadata |  emd-25712.cif.gz emd-25712.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25712 http://ftp.pdbj.org/pub/emdb/structures/EMD-25712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25712 | HTTPS FTP |
-Related structure data
| Related structure data |  7t6bMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25712.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25712.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.842 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Structure of S1PR2-heterotrimeric G13 signaling complex
| Entire | Name: Structure of S1PR2-heterotrimeric G13 signaling complex |
|---|---|
| Components |
|
-Supramolecule #1: Structure of S1PR2-heterotrimeric G13 signaling complex
| Supramolecule | Name: Structure of S1PR2-heterotrimeric G13 signaling complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein subunit alpha-13
| Macromolecule | Name: Guanine nucleotide-binding protein subunit alpha-13 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.255273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA ARLVKILLLG AGESGKSTFL KQMRIIHGQD FDQRAREEFR PTIYSNVIKG MRVLVDARE KLHIPWGDNS NQQHGDKMMS FDTRAPMAAQ GMVETRVFLQ YLPAIRALWA DSGIQNAYDR RREFQLGESV K YFLDNLDK ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA ARLVKILLLG AGESGKSTFL KQMRIIHGQD FDQRAREEFR PTIYSNVIKG MRVLVDARE KLHIPWGDNS NQQHGDKMMS FDTRAPMAAQ GMVETRVFLQ YLPAIRALWA DSGIQNAYDR RREFQLGESV K YFLDNLDK LGEPDYIPSQ QDILLARRPT KGIHEYDFEI KNVPFKMVDV GGQRSERKRW FECFDSVTSI LFLVSSSEFD QV LMEDRLT NRLTESLNIF ETIVNNRVFS NVSIILFLNK TDLLEEKVQV VSIKDYFLEF EGDPHCLRDV QKFLVECFRG KRR DQQQKP LYHHFTTAIN TENIRLVFRD VKDTILHDNL KQLMLQ UniProtKB: Guanine nucleotide-binding protein subunit alpha-13 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.97066 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HLEVLFQGPG SSGSELDQLR QEAEQLKNQI RDARKACADA TLSQITNNID PVGRIQMRTR RTLRGHLAKI YAMHWGTDS RLLVSASQDG KLIIWDSYTT NKVHAIPLRS SWVMTCAYAP SGNYVACGGL DNICSIYNLK TREGNVRVSR E LAGHTGYL ...String: MHHHHHHHHH HLEVLFQGPG SSGSELDQLR QEAEQLKNQI RDARKACADA TLSQITNNID PVGRIQMRTR RTLRGHLAKI YAMHWGTDS RLLVSASQDG KLIIWDSYTT NKVHAIPLRS SWVMTCAYAP SGNYVACGGL DNICSIYNLK TREGNVRVSR E LAGHTGYL SCCRFLDDNQ IVTSSGDTTC ALWDIETGQQ TTTFTGHTGD VMSLSLAPDT RLFVSGACDA SAKLWDVREG MC RQTFTGH ESDINAICFF PNGNAFATGS DDATCRLFDL RADQELMTYS HDNIICGITS VSFSKSGRLL LAGYDDFNCN VWD ALKADR AGVLAGHDNR VSCLGVTDDG MAVATGSWDS FLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.784896 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAHHHHHH HH |
-Macromolecule #5: Sphingosine 1-phosphate receptor 2
| Macromolecule | Name: Sphingosine 1-phosphate receptor 2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.902824 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGSLYSEYLN PNKVQEHYNY TKETLETQET TSRQVASAFI VILCCAIVVE NLLVLIAVAR NSKFHSAMYL FLGNLAASDL LAGVAFVAN TLLSGSVTLR LTPVQWFARE GSAFITLSAS VFSLLAIAIE RHVAIAKVKL YGSDKSCRML LLIGASWLIS L VLGGLPIL ...String: MGSLYSEYLN PNKVQEHYNY TKETLETQET TSRQVASAFI VILCCAIVVE NLLVLIAVAR NSKFHSAMYL FLGNLAASDL LAGVAFVAN TLLSGSVTLR LTPVQWFARE GSAFITLSAS VFSLLAIAIE RHVAIAKVKL YGSDKSCRML LLIGASWLIS L VLGGLPIL GWNCLGHLEA CSTVLPLYAK HYVLCVVTIF SIILLAIVAL YVRIYCVVRS SHADMAAPQT LALLKTVTIV LG VFIVCWL PAFSILLLDY ACPVHSCPIL YKAHYFFAVS TLNSLLNPVI YTWRSRDLRR EVLRPLQCWR PGVGVQGRRR GGT PGHHLL PLRSSSSLER GMHMPTSPTF LEGNTVVDYK DDDDK UniProtKB: Sphingosine 1-phosphate receptor 2 |
-Macromolecule #6: (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate
| Macromolecule | Name: (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate type: ligand / ID: 6 / Number of copies: 1 / Formula: S1P |
|---|---|
| Molecular weight | Theoretical: 379.472 Da |
| Chemical component information | 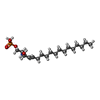 ChemComp-S1P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 20 mM Hepes pH7.5, 150 mM NaCl, 0.001% L-MNG/0.0001% CHS, 0.0025% GDN | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















