[English] 日本語
 Yorodumi
Yorodumi- EMDB-24830: CryoEM structure of Methylotuvimicrobium alcaliphilum 20Z pMMO in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
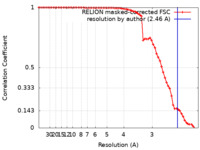
| Title | CryoEM structure of Methylotuvimicrobium alcaliphilum 20Z pMMO in a POPC nanodisc at 2.46 Angstrom resolution | |||||||||
 Map data Map data | CryoEM structure of Methylotuvimicrobium alcaliphilum 20Z in a POPC nanodisc at 2.42 angstrom resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmethane monooxygenase (soluble) / methane monooxygenase [NAD(P)H] activity / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Methylotuvimicrobium alcaliphilum 20Z (bacteria) / Methylotuvimicrobium alcaliphilum 20Z (bacteria) /  Methylomicrobium alcaliphilum (strain DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z) (bacteria) Methylomicrobium alcaliphilum (strain DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.46 Å | |||||||||
 Authors Authors | Koo CW / Rosenzweig AC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer. Authors: Christopher W Koo / Frank J Tucci / Yuan He / Amy C Rosenzweig /  Abstract: Bacterial methane oxidation using the enzyme particulate methane monooxygenase (pMMO) contributes to the removal of environmental methane, a potent greenhouse gas. Crystal structures determined using ...Bacterial methane oxidation using the enzyme particulate methane monooxygenase (pMMO) contributes to the removal of environmental methane, a potent greenhouse gas. Crystal structures determined using inactive, detergent-solubilized pMMO lack several conserved regions neighboring the proposed active site. We show that reconstituting pMMO in nanodiscs with lipids extracted from the native organism restores methane oxidation activity. Multiple nanodisc-embedded pMMO structures determined by cryo-electron microscopy to 2.14- to 2.46-angstrom resolution reveal the structure of pMMO in a lipid environment. The resulting model includes stabilizing lipids, regions of the PmoA and PmoC subunits not observed in prior structures, and a previously undetected copper-binding site in the PmoC subunit with an adjacent hydrophobic cavity. These structures provide a revised framework for understanding and engineering pMMO function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24830.map.gz emd_24830.map.gz | 195.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24830-v30.xml emd-24830-v30.xml emd-24830.xml emd-24830.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24830_fsc.xml emd_24830_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_24830.png emd_24830.png | 122.1 KB | ||
| Filedesc metadata |  emd-24830.cif.gz emd-24830.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24830 http://ftp.pdbj.org/pub/emdb/structures/EMD-24830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24830 | HTTPS FTP |
-Related structure data
| Related structure data |  7s4lMC  7s4hC  7s4iC  7s4jC  7s4kC  7s4mC  7t4oC  7t4pC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24830.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24830.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of Methylotuvimicrobium alcaliphilum 20Z in a POPC nanodisc at 2.42 angstrom resolution | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.525 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : pMMO complex in a POPC nanodisc
| Entire | Name: pMMO complex in a POPC nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: pMMO complex in a POPC nanodisc
| Supramolecule | Name: pMMO complex in a POPC nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Methylotuvimicrobium alcaliphilum 20Z (bacteria) Methylotuvimicrobium alcaliphilum 20Z (bacteria) |
-Macromolecule #1: Particulate methane monooxygenase, B subunit
| Macromolecule | Name: Particulate methane monooxygenase, B subunit / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: methane monooxygenase (soluble) |
|---|---|
| Source (natural) | Organism:  Methylomicrobium alcaliphilum (strain DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z) (bacteria) Methylomicrobium alcaliphilum (strain DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z) (bacteria)Strain: DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z |
| Molecular weight | Theoretical: 45.602125 KDa |
| Sequence | String: MKIIKDKVAK LSFVALLVTV TAAMFYTPTA SAHGEKSQAA FMRMRTIHWF DLNWSKDQVS VNETMSISGK FHVFAGWPET VDKPEVAFL NIGIPGPVFI RAGSWIGGQL VPRSVSLELG ETYEFKVLLK ARRPGDWHVH TMMNVQGGGP IIGPGKWVTI T GSMGDFKN ...String: MKIIKDKVAK LSFVALLVTV TAAMFYTPTA SAHGEKSQAA FMRMRTIHWF DLNWSKDQVS VNETMSISGK FHVFAGWPET VDKPEVAFL NIGIPGPVFI RAGSWIGGQL VPRSVSLELG ETYEFKVLLK ARRPGDWHVH TMMNVQGGGP IIGPGKWVTI T GSMGDFKN PITTLTGETI DLETYALDGV YGWHLFWYLL GVAWMVYWCR KPVFIPRRIA VDAGKADSLI TPTDKKVGMA FA AGTLAIV AVSMGQANEK YPVTTPLQAG LMRGIKSLEL PQPTVSVKVV DASYRVPGRA MQMTLEITNN GDSAVRLAEF NTA SVRFLD ADVYEDDTNY PDDLLAEEGL SVSDNSPLAP GETRTVDVTA SDAAWEVYRL ADLIYDPDSR FAGLLFFIDE DGNR QMTMV DAPLIPTFI UniProtKB: Particulate methane monooxygenase, B subunit |
-Macromolecule #2: Particulate methane monooxygenase, A subunit
| Macromolecule | Name: Particulate methane monooxygenase, A subunit / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: methane monooxygenase (soluble) |
|---|---|
| Source (natural) | Organism:  Methylomicrobium alcaliphilum (strain DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z) (bacteria) Methylomicrobium alcaliphilum (strain DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z) (bacteria)Strain: DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z |
| Molecular weight | Theoretical: 28.277934 KDa |
| Sequence | String: MSASQSAVRS RAEAVKVSRT FDYMILFTVF FVVLGGYHIH YMLTGGDWDF WTDWKDRRLW VTVAPIVSIT FPAAVQAVLW WRYRIAWGA TLCVLGLLLG EWINRYFNFW GWTYFPVNFV FPSNLMPGAI VLDVILMLSN SMTLTAVVGG LAWGLLFYPG N WPIIAPLH ...String: MSASQSAVRS RAEAVKVSRT FDYMILFTVF FVVLGGYHIH YMLTGGDWDF WTDWKDRRLW VTVAPIVSIT FPAAVQAVLW WRYRIAWGA TLCVLGLLLG EWINRYFNFW GWTYFPVNFV FPSNLMPGAI VLDVILMLSN SMTLTAVVGG LAWGLLFYPG N WPIIAPLH VPVEYNGMMM TLADLQGYHY VRTGTPEYIR MVEKGTLRTF GKDVAPVSAF FSGFVSILIY FLWHFFGSWF GS EKFVQAA UniProtKB: Particulate methane monooxygenase, A subunit |
-Macromolecule #3: Particulate methane monooxygenase, C subunit
| Macromolecule | Name: Particulate methane monooxygenase, C subunit / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO / EC number: methane monooxygenase (soluble) |
|---|---|
| Source (natural) | Organism:  Methylomicrobium alcaliphilum (strain DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z) (bacteria) Methylomicrobium alcaliphilum (strain DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z) (bacteria)Strain: DSM 19304 / NCIMB 14124 / VKM B-2133 / 20Z |
| Molecular weight | Theoretical: 28.872328 KDa |
| Sequence | String: MAATTESVKA DAAEAPLLNK KNIIAGASLY LVFYAWVRWY EGVYGWSAGL DSFAPEFETY WMNFLYIEMV LEVLVASVLW GYIWKSRDR KVMSITPREE LRRHFTHWTW LMMYGIAIYF GASYFTEQDG TWHQTIVRDT DFTPSHIIEF YLSYPIYIIT G GASFLYAK ...String: MAATTESVKA DAAEAPLLNK KNIIAGASLY LVFYAWVRWY EGVYGWSAGL DSFAPEFETY WMNFLYIEMV LEVLVASVLW GYIWKSRDR KVMSITPREE LRRHFTHWTW LMMYGIAIYF GASYFTEQDG TWHQTIVRDT DFTPSHIIEF YLSYPIYIIT G GASFLYAK TRLPTYQQGL SLQYLVVVVG PFMILPNVGL NEWGHTFWFM EELFVAPLHY GFVFFGWSAL GVLGVINIEL GA LSKLLKK DLA UniProtKB: Particulate methane monooxygenase, C subunit |
-Macromolecule #4: COPPER (II) ION
| Macromolecule | Name: COPPER (II) ION / type: ligand / ID: 4 / Number of copies: 9 / Formula: CU |
|---|---|
| Molecular weight | Theoretical: 63.546 Da |
| Chemical component information |  ChemComp-CU: |
-Macromolecule #5: 1,2-dihexanoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1,2-dihexanoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 5 / Number of copies: 15 / Formula: HXG |
|---|---|
| Molecular weight | Theoretical: 454.515 Da |
| Chemical component information | 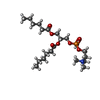 ChemComp-HXG: |
-Macromolecule #6: DECANE
| Macromolecule | Name: DECANE / type: ligand / ID: 6 / Number of copies: 9 / Formula: D10 |
|---|---|
| Molecular weight | Theoretical: 142.282 Da |
| Chemical component information |  ChemComp-D10: |
-Macromolecule #7: (S)-2,3-bis(hexanoyloxy)propyl(2-(trimethylammonio)ethyl)phosphate
| Macromolecule | Name: (S)-2,3-bis(hexanoyloxy)propyl(2-(trimethylammonio)ethyl)phosphate type: ligand / ID: 7 / Number of copies: 3 / Formula: 6ER |
|---|---|
| Molecular weight | Theoretical: 454.515 Da |
| Chemical component information | 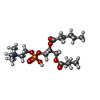 ChemComp-6ER: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 3 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.3 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)