+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24732 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Multidrug Efflux pump AdeJ with TP-6076 bound | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | multidrug efflux pump / MEMBRANE PROTEIN-ANTIBIOTIC complex | |||||||||
| Function / homology | Hydrophobe/amphiphile efflux-1 HAE1 / Multidrug efflux transporter AcrB TolC docking domain, DN/DC subdomains / Acriflavin resistance protein / AcrB/AcrD/AcrF family / efflux transmembrane transporter activity / xenobiotic transmembrane transporter activity / response to toxic substance / plasma membrane / Efflux pump membrane transporter Function and homology information Function and homology information | |||||||||
| Biological species |  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Zhang Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2021 Journal: mBio / Year: 2021Title: An Analysis of the Novel Fluorocycline TP-6076 Bound to Both the Ribosome and Multidrug Efflux Pump AdeJ from Acinetobacter baumannii. Authors: Christopher E Morgan / Zhemin Zhang / Robert A Bonomo / Edward W Yu /  Abstract: Antibiotic resistance among bacterial pathogens continues to pose a serious global health threat. Multidrug-resistant (MDR) strains of the Gram-negative organism Acinetobacter baumannii utilize a ...Antibiotic resistance among bacterial pathogens continues to pose a serious global health threat. Multidrug-resistant (MDR) strains of the Gram-negative organism Acinetobacter baumannii utilize a number of resistance determinants to evade current antibiotics. One of the major resistance mechanisms employed by these pathogens is the use of multidrug efflux pumps. These pumps extrude xenobiotics directly out of bacterial cells, resulting in treatment failures when common antibiotics are administered. Here, the structure of the novel tetracycline antibiotic TP-6076, bound to both the cinetobacter rug fflux pump AdeJ and the ribosome from Acinetobacter baumannii, using single-particle cryo-electron microscopy (cryo-EM), is elucidated. In this work, the structure of the AdeJ-TP-6076 complex is solved, and we show that AdeJ utilizes a network of hydrophobic interactions to recognize this fluorocycline. Concomitant with this, we elucidate three structures of TP-6076 bound to the A. baumannii ribosome and determine that its binding is stabilized largely by electrostatic interactions. We then compare the differences in binding modes between TP-6076 and the related tetracycline antibiotic eravacycline in both targets. These differences suggest that modifications to the tetracycline core may be able to alter AdeJ binding while maintaining interactions with the ribosome. Together, this work highlights how different mechanisms are used to stabilize the binding of tetracycline-based compounds to unique bacterial targets and provides guidance for the future clinical development of tetracycline antibiotics. Treatment of antibiotic-resistant organisms such as A. baumannii represents an ongoing issue for modern medicine. The multidrug efflux pump AdeJ serves as a major resistance determinant in A. baumannii through its action of extruding antibiotics from the cell. In this work, we use cryo-EM to show how AdeJ recognizes the experimental tetracycline antibiotic TP-6076 and prevents this drug from interacting with the A. baumannii ribosome. Since AdeJ and the ribosome use different binding modes to stabilize interactions with TP-6076, exploiting these differences may guide future drug development for combating antibiotic-resistant A. baumannii and potentially other strains of MDR bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24732.map.gz emd_24732.map.gz | 8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24732-v30.xml emd-24732-v30.xml emd-24732.xml emd-24732.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24732.png emd_24732.png | 191.8 KB | ||
| Filedesc metadata |  emd-24732.cif.gz emd-24732.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24732 http://ftp.pdbj.org/pub/emdb/structures/EMD-24732 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24732 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24732 | HTTPS FTP |
-Validation report
| Summary document |  emd_24732_validation.pdf.gz emd_24732_validation.pdf.gz | 555.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24732_full_validation.pdf.gz emd_24732_full_validation.pdf.gz | 555.1 KB | Display | |
| Data in XML |  emd_24732_validation.xml.gz emd_24732_validation.xml.gz | 4.4 KB | Display | |
| Data in CIF |  emd_24732_validation.cif.gz emd_24732_validation.cif.gz | 4.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24732 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24732 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24732 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24732 | HTTPS FTP |
-Related structure data
| Related structure data |  7ry3MC  7ryfC  7rygC  7ryhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24732.map.gz / Format: CCP4 / Size: 8.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24732.map.gz / Format: CCP4 / Size: 8.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : multidrug efflux pump AdeJ
| Entire | Name: multidrug efflux pump AdeJ |
|---|---|
| Components |
|
-Supramolecule #1: multidrug efflux pump AdeJ
| Supramolecule | Name: multidrug efflux pump AdeJ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
-Macromolecule #1: Efflux pump membrane transporter
| Macromolecule | Name: Efflux pump membrane transporter / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 114.637781 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAQFFIHRPI FAWVIALVIM LAGILTLTKM PIAQYPTIAP PTVTIAATYP GASAETVENT VTQIIEQQMN GLDGLRYISS NSAGNGQAS IQLNFEQGVD PDIAQVQVQN KLQSATALLP EDVQRQGVTV TKSGASFLQV IAFYSPDNNL SDSDIKDYVN S SIKEPLSR ...String: MAQFFIHRPI FAWVIALVIM LAGILTLTKM PIAQYPTIAP PTVTIAATYP GASAETVENT VTQIIEQQMN GLDGLRYISS NSAGNGQAS IQLNFEQGVD PDIAQVQVQN KLQSATALLP EDVQRQGVTV TKSGASFLQV IAFYSPDNNL SDSDIKDYVN S SIKEPLSR VAGVGEVQVF GGSYAMRIWL DPAKLTSYQL TPSDIATALQ AQNSQVAVGQ LGGAPAVQGQ VLNATVNAQS LL QTPEQFK NIFLKNTASG AEVRLKDVAR VELGSDNYQF DSKFNGKPAA GLAIKIATGA NALDTAEAVE QRLSELRKNY PTG LADKLA YDTTPFIRLS IESVVHTLIE AVILVFIVMF LFLQNWRATI IPTLAVPVVV LGTFAVINIF GFSINTLTMF AMVL AIGLL VDDAIVVVEN VERVMSEDHT DPVTATSRSM QQISGALVGI TSVLTAVFVP MAFFGGTTGV IYRQFSITLV TAMVL SLIV ALTFTPALCA TILKQHDPNK EPSNNIFARF FRSFNNGFDR MSHSYQNGVS RMLKGKIFSG VLYAVVVALL VFLFQK LPS SFLPEEDQGV VMTLVQLPPN ATLDRTGKVI DTMTNFFMNE KDTVESIFTV SGFSFTGVGQ NAGIGFVKLK DWSKRTT PE TQIGSLIQRG MALNMIIKDA SYVMPLQLPA MPELGVTAGF NLQLKDSSGQ GHEKLIAARN TILGLASQDK RLVGVRPN G QEDTPQYQIN VDQAQAGAMG VSIAEINNTM RIAWGGSYIN DFVDRGRVKK VYVQGDAGSR MMPEDLNKWY VRNNKGEMV PFSAFATGEW TYGSPRLERY NGVSSVNIQG TPAPGVSSGD AMKAMEEIIG KLPSMGLQGF DYEWTGLSLE ERESGAQAPF LYALSLLIV FLCLAALYES WSIPFSVLLV VPLGVIGAIV LTYLGMIIKG DPNLSNNIYF QVAIIAVIGL SAKNAILIVE F AKELQEKG EDLLDATLHA AKMRLRPIIM TTLAFGFGVL PLALSTGAGA GSQHSVGFGV LGGVLSATFL GIFFIPVFYV WI RSIFKYK PKTINTQEHK S UniProtKB: Efflux pump membrane transporter |
-Macromolecule #2: (4S,4aS,5aR,12aS)-4-(diethylamino)-3,10,12,12a-tetrahydroxy-1,11-...
| Macromolecule | Name: (4S,4aS,5aR,12aS)-4-(diethylamino)-3,10,12,12a-tetrahydroxy-1,11-dioxo-8-[(2S)-pyrrolidin-2-yl]-7-(trifluoromethyl)-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide type: ligand / ID: 2 / Number of copies: 1 / Formula: 80P |
|---|---|
| Molecular weight | Theoretical: 579.565 Da |
| Chemical component information | 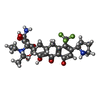 ChemComp-80P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 36.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.91 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 68346 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)