[English] 日本語
 Yorodumi
Yorodumi- EMDB-24484: Complex III2 from Candida albicans, inhibitor free, Rieske head d... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24484 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex III2 from Candida albicans, inhibitor free, Rieske head domain in intermediate position | ||||||||||||||||||
 Map data Map data | Candida albicans complex III2, inhibitor free, Rieske head domain intermediate state | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of filamentous growth / matrix side of mitochondrial inner membrane / protein processing involved in protein targeting to mitochondrion / mitochondrial respiratory chain complex III assembly / : / quinol-cytochrome-c reductase / ubiquinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / mitochondrial crista / heterochromatin ...regulation of filamentous growth / matrix side of mitochondrial inner membrane / protein processing involved in protein targeting to mitochondrion / mitochondrial respiratory chain complex III assembly / : / quinol-cytochrome-c reductase / ubiquinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / mitochondrial crista / heterochromatin / aerobic respiration / metalloendopeptidase activity / 2 iron, 2 sulfur cluster binding / electron transfer activity / oxidoreductase activity / heme binding / mitochondrion / membrane / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Candida albicans SC5314 (yeast) / Candida albicans SC5314 (yeast) /  Yeast (yeast) Yeast (yeast) | ||||||||||||||||||
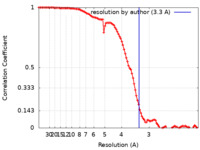
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||
 Authors Authors | Di Trani JM / Rubinstein JL | ||||||||||||||||||
| Funding support |  Canada, Canada,  Sweden, 5 items Sweden, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III. Authors: Justin M Di Trani / Zhongle Liu / Luke Whitesell / Peter Brzezinski / Leah E Cowen / John L Rubinstein /   Abstract: Electron transfer between respiratory complexes drives transmembrane proton translocation, which powers ATP synthesis and membrane transport. The homodimeric respiratory complex III (CIII) oxidizes ...Electron transfer between respiratory complexes drives transmembrane proton translocation, which powers ATP synthesis and membrane transport. The homodimeric respiratory complex III (CIII) oxidizes ubiquinol to ubiquinone, transferring electrons to cytochrome c and translocating protons through a mechanism known as the Q cycle. The Q cycle involves ubiquinol oxidation and ubiquinone reduction at two different sites within each CIII monomer, as well as movement of the head domain of the Rieske subunit. We determined structures of Candida albicans CIII by cryoelectron microscopy (cryo-EM), revealing endogenous ubiquinone and visualizing the continuum of Rieske head domain conformations. Analysis of these conformations does not indicate cooperativity in the Rieske head domain position or ligand binding in the two CIIIs of the CIII dimer. Cryo-EM with the indazole derivative Inz-5, which inhibits fungal CIII and is fungicidal when administered with fungistatic azole drugs, showed that Inz-5 inhibition alters the equilibrium of Rieske head domain positions. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24484.map.gz emd_24484.map.gz | 2.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24484-v30.xml emd-24484-v30.xml emd-24484.xml emd-24484.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24484_fsc.xml emd_24484_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_24484.png emd_24484.png | 76.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24484 http://ftp.pdbj.org/pub/emdb/structures/EMD-24484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24484 | HTTPS FTP |
-Validation report
| Summary document |  emd_24484_validation.pdf.gz emd_24484_validation.pdf.gz | 347.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24484_full_validation.pdf.gz emd_24484_full_validation.pdf.gz | 347.2 KB | Display | |
| Data in XML |  emd_24484_validation.xml.gz emd_24484_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_24484_validation.cif.gz emd_24484_validation.cif.gz | 15.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24484 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24484 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24484 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24484 | HTTPS FTP |
-Related structure data
| Related structure data |  7rjcMC  7rjaC  7rjbC  7rjdC  7rjeC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24484.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24484.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Candida albicans complex III2, inhibitor free, Rieske head domain intermediate state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Respiratory complex III2 from Candida albicans
+Supramolecule #1: Respiratory complex III2 from Candida albicans
+Macromolecule #1: Ubiquinol--cytochrome-c reductase subunit
+Macromolecule #2: Cytochrome b
+Macromolecule #3: Ubiquinol--cytochrome-c reductase catalytic subunit
+Macromolecule #4: Cytochrome b-c1 complex subunit 7
+Macromolecule #5: Ubiquinol--cytochrome-c reductase subunit 8
+Macromolecule #6: Ubiquinol--cytochrome-c reductase subunit 6
+Macromolecule #7: Ubiquinol--cytochrome-c reductase subunit 9
+Macromolecule #8: Cytochrome b-c1 complex subunit 2, mitochondrial
+Macromolecule #9: Cytochrome b-c1 complex subunit Rieske, mitochondrial
+Macromolecule #10: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #11: UBIQUINONE-10
+Macromolecule #12: HEME C
+Macromolecule #13: FE2/S2 (INORGANIC) CLUSTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)