[English] 日本語
 Yorodumi
Yorodumi- EMDB-23635: Engineered disulfide cross-linked closed conformation of the Yeas... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23635 | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
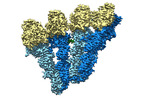
| Title | Engineered disulfide cross-linked closed conformation of the Yeast gamma-TuRC(SS) | ||||||||||||||||||||||||||||||||||||||||||
 Map data Map data | Unmasked unsharpened gamma-TuRC(SS) main map | ||||||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | microtubule nucleation / CELL CYCLE | ||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationgamma-tubulin complex localization to nuclear side of mitotic spindle pole body / protein localization to mitotic spindle pole body / inner plaque of spindle pole body / microtubule nucleation by spindle pole body / outer plaque of spindle pole body / gamma-tubulin small complex / central plaque of spindle pole body / karyogamy involved in conjugation with cellular fusion / regulation of microtubule nucleation / microtubule nucleator activity ...gamma-tubulin complex localization to nuclear side of mitotic spindle pole body / protein localization to mitotic spindle pole body / inner plaque of spindle pole body / microtubule nucleation by spindle pole body / outer plaque of spindle pole body / gamma-tubulin small complex / central plaque of spindle pole body / karyogamy involved in conjugation with cellular fusion / regulation of microtubule nucleation / microtubule nucleator activity / mitotic spindle pole body / mitotic spindle elongation / gamma-tubulin complex / gamma-tubulin ring complex / meiotic spindle organization / positive regulation of microtubule nucleation / microtubule nucleation / spindle pole body / gamma-tubulin binding / positive regulation of cytoplasmic translation / mitotic sister chromatid segregation / spindle assembly / cytoplasmic microtubule organization / mitotic spindle organization / meiotic cell cycle / structural constituent of cytoskeleton / spindle / spindle pole / mitotic cell cycle / protein-containing complex assembly / microtubule / cytoskeleton / calmodulin binding / GTP binding / protein-containing complex binding / nucleus / cytoplasm Similarity search - Function | ||||||||||||||||||||||||||||||||||||||||||
| Biological species |   | ||||||||||||||||||||||||||||||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Brilot AF / Lyon AS | ||||||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, 13 items United States, 13 items
| ||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: CM1-driven assembly and activation of yeast γ-tubulin small complex underlies microtubule nucleation. Authors: Axel F Brilot / Andrew S Lyon / Alex Zelter / Shruthi Viswanath / Alison Maxwell / Michael J MacCoss / Eric G Muller / Andrej Sali / Trisha N Davis / David A Agard /  Abstract: Microtubule (MT) nucleation is regulated by the γ-tubulin ring complex (γTuRC), conserved from yeast to humans. In , γTuRC is composed of seven identical γ-tubulin small complex (γTuSC) sub- ...Microtubule (MT) nucleation is regulated by the γ-tubulin ring complex (γTuRC), conserved from yeast to humans. In , γTuRC is composed of seven identical γ-tubulin small complex (γTuSC) sub-assemblies, which associate helically to template MT growth. γTuRC assembly provides a key point of regulation for the MT cytoskeleton. Here, we combine crosslinking mass spectrometry, X-ray crystallography, and cryo-EM structures of both monomeric and dimeric γTuSCs, and open and closed helical γTuRC assemblies in complex with Spc110p to elucidate the mechanisms of γTuRC assembly. γTuRC assembly is substantially aided by the evolutionarily conserved CM1 motif in Spc110p spanning a pair of adjacent γTuSCs. By providing the highest resolution and most complete views of any γTuSC assembly, our structures allow phosphorylation sites to be mapped, surprisingly suggesting that they are mostly inhibitory. A comparison of our structures with the CM1 binding site in the human γTuRC structure at the interface between GCP2 and GCP6 allows for the interpretation of significant structural changes arising from CM1 helix binding to metazoan γTuRC. | ||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23635.map.gz emd_23635.map.gz | 764.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23635-v30.xml emd-23635-v30.xml emd-23635.xml emd-23635.xml | 28.5 KB 28.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23635.png emd_23635.png | 230 KB | ||
| Filedesc metadata |  emd-23635.cif.gz emd-23635.cif.gz | 8.3 KB | ||
| Others |  emd_23635_half_map_1.map.gz emd_23635_half_map_1.map.gz emd_23635_half_map_2.map.gz emd_23635_half_map_2.map.gz | 160.2 MB 160.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23635 http://ftp.pdbj.org/pub/emdb/structures/EMD-23635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23635 | HTTPS FTP |
-Related structure data
| Related structure data |  7m2wMC  7m2xC  7m2yC  7m2zC  7m3pC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23635.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23635.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked unsharpened gamma-TuRC(SS) main map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: gamma-TuRC(SS) half map 2
| File | emd_23635_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | gamma-TuRC(SS) half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: gamma-TuRC(SS) half map 1
| File | emd_23635_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | gamma-TuRC(SS) half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Engineered disulfide cross-linked closed conformation of the Yeas...
| Entire | Name: Engineered disulfide cross-linked closed conformation of the Yeast gamma-TuRC(SS) |
|---|---|
| Components |
|
-Supramolecule #1: Engineered disulfide cross-linked closed conformation of the Yeas...
| Supramolecule | Name: Engineered disulfide cross-linked closed conformation of the Yeast gamma-TuRC(SS) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin gamma chain
| Macromolecule | Name: Tubulin gamma chain / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 52.73334 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGGEIITLQA GQCGNHVGKF LWSQLAKEHA IGTDGLSQLP DSSTERDDDT KPFFRENCRN KFTPRAIMMD SEPSVIADVE NTFRGFFDP RNTWVASDGA SAGNSWANGY DIGTRNQDDI LNKIDKEIDS TDNFEGFQLL HSVAGGTGSG LGSNLLEALC D RYPKKILT ...String: MGGEIITLQA GQCGNHVGKF LWSQLAKEHA IGTDGLSQLP DSSTERDDDT KPFFRENCRN KFTPRAIMMD SEPSVIADVE NTFRGFFDP RNTWVASDGA SAGNSWANGY DIGTRNQDDI LNKIDKEIDS TDNFEGFQLL HSVAGGTGSG LGSNLLEALC D RYPKKILT TYSVFPARSS EVVVQSYNTI LALRRLIEDS DATVVFDNAS LLNISGKVFR NPNIDLQHTN QLISTIISSV TN SIRFPSY MYSSMSSIYS TLIPSPELHF LSPSFTPFTS DYIHDDIAHK CHSSYDVMLD LLDPSNSLVS TAMNNPTYFN VYN TIIGNV EPRQISRAMT KLQQRIKFPS WSSSAMHVNI GRRSPYLPLQ PNENEVSGMM LSNMSTVVNV FENACNTFDK VFAK GAFLN NYNVGDLFQS MQNVQDEFAE SREVVQSLME DYVAAEQDSY LDDVLVDDEN MVGELEEDLD ADGDHKLV UniProtKB: Tubulin gamma chain |
-Macromolecule #2: Spindle pole body component SPC97
| Macromolecule | Name: Spindle pole body component SPC97 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 96.940594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEIKEVDDRA ELLRYTNNIP LLGKLVNHQP LWSTNPKLKS FSLEKISAPD QRRVQEALVV KDLLNVLIGL EGTYIRYFND YEPSDPETP IEFKIAKKMD PSFKTFSRRI VRYGKQYMIL TRAYEKWSDT SFGMVLQRFA YEIRRFLEDV YLKTLVERLE R DFNKVPNF ...String: MEIKEVDDRA ELLRYTNNIP LLGKLVNHQP LWSTNPKLKS FSLEKISAPD QRRVQEALVV KDLLNVLIGL EGTYIRYFND YEPSDPETP IEFKIAKKMD PSFKTFSRRI VRYGKQYMIL TRAYEKWSDT SFGMVLQRFA YEIRRFLEDV YLKTLVERLE R DFNKVPNF SIRELEQIIN ETEVNKQMEL LYNIYEEIFR EIEERRTNQS SQEDFNNFMD SMKNESSLHL RLMVAFDTTV YP VPKGGAI LKIFQQKILE NLGDRSSVMF LKKLLNNISQ DYCTMLYEWL TQGILNDPYQ EFMTYDDLEG KTDNIFDTRD RAW DTQYFI RKDVLLRDCD SEEDKNLLFK MLRTGILLKV VRASLQIPTI PSNSSDITIQ EINDFADLME GSNLELYVDK CYSR ANEIF LKLFFQGYDL INVLKHLQQI FLGYQSGHNV LKFLTKNMGE LTKHYRNDNN ANYDKLLQNF ELERQSENPN NLMRQ LLMI QFDTETLPQV LSHYLQIYPE VPENNSANDD SDPLMHANNF KNMNAILFDE LSKERTGAYH GSNLELYTPK SAIYHL KFD INIPYPLNII ISRTCMIKYQ IILRYQLVLQ YHSRLLDETW MDLNKTPSWK YRGYSHTVKR RIVRATRVLH AKMNHFI KT IMEYFNQNVI DKEVYSLEKC YRNPTLAVAI QNELEGGLTN IMTNRCLSDL IPLQLQIFDI VYKFCKFIKS MRAKLCQL D PVLYEKHKSG MMKTLNEGYR TNNGGQEDVG YQEDAALELI QKLIEYISNA SSIFRKCLIN FTQELSTEKF DFYDSSSVD AAGIERVLYS IVPPRSASAS SQR UniProtKB: Spindle pole body component SPC97 |
-Macromolecule #3: Spindle pole body component SPC98
| Macromolecule | Name: Spindle pole body component SPC98 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 98.336211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MELEPTLFGI IEALAPQLLS QSHLQTFVSD VVNLLRSSTK SATQLGPLID FYKLQSLDSP ETTIMWHKIE KFLDALFGIQ NTDDMVKYL SVFQSLLPSN YRAKIVQKSS GLNMENLANH EHLLSPVRAP SIYTEASFEN MDRFSERRSM VSSPNRYVPS S TYSSVTLR ...String: MELEPTLFGI IEALAPQLLS QSHLQTFVSD VVNLLRSSTK SATQLGPLID FYKLQSLDSP ETTIMWHKIE KFLDALFGIQ NTDDMVKYL SVFQSLLPSN YRAKIVQKSS GLNMENLANH EHLLSPVRAP SIYTEASFEN MDRFSERRSM VSSPNRYVPS S TYSSVTLR QLSNPYYVNT IPEEDILKYV SYTLLATTSA LFPFDHEQIQ IPSKIPNFES GLLHLIFEAG LLYQSLGYKV EK FRMLNIS PMKKALIIEI SEELQNYTAF VNNLVSSGTV VSLKSLYREI YENIIRLRIY CRFTEHLEEL SGDTFLIELN IFK SHGDLT IRKIATNLFN SMISLYYEYL MNWLTKGLLR ATYGEFFIAE NTDTNGTDDD FIYHIPIEFN QERVPAFIPK ELAY KIFMI GKSYIFLEKY CKEVQWTNEF SKKYHVLYQS NSYRGISTNF FEIINDQYSE IVNHTNQILN QKFHYRDVVF ALKNI LLMG KSDFMDALIE KANDILATPS DSLPNYKLTR VLQEAVQLSS LRHLMNSPRN SSVINGLDAR VLDLGHGSVG WDVFTL DYI LYPPLSLVLN VNRPFGRKEY LRIFNFLWRF KKNNYFYQKE MLKSNDIIRS FKKIRGYNPL IRDIINKLSR ISILRTQ FQ QFNSKMESYY LNCIIEENFK EMTRKLQRTE NKSQNQFDLI RLNNGTIELN GILTPKAEVL TKSSSSKPQK HAIEKTLN I DELESVHNTF LTNILSHKLF ATNTSEISVG DYSGQPYPTS LVLLLNSVYE FVKVYCNLND IGYEIFIKMN LNDHEASNG LLGKFNTNLK EIVSQYKNFK DRLYIFRADL KNDGDEELFL LSKSLR UniProtKB: Spindle pole body component SPC98 |
-Macromolecule #4: Spindle pole body component 110
| Macromolecule | Name: Spindle pole body component 110 / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 25.449555 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDEASHLPNG SLKNMEFTPV GFIKSKRNTT QTQVVSPTKV PNANNGDENE GPVKKRQRRS IDDTIDSTRL FSEASQFDDS FPEIKANIP PSPRSGNVDK SRKRNLIDDL KKDVPMSQPL KEQEVREHQM KKERFDRALE SKLLGKRHIT YANSDISNKE L YINEIKSL ...String: MDEASHLPNG SLKNMEFTPV GFIKSKRNTT QTQVVSPTKV PNANNGDENE GPVKKRQRRS IDDTIDSTRL FSEASQFDDS FPEIKANIP PSPRSGNVDK SRKRNLIDDL KKDVPMSQPL KEQEVREHQM KKERFDRALE SKLLGKRHIT YANSDISNKE L YINEIKSL KHEIKELRKE KNDTLNNYDT LEEETDDLKN RLQALEKELD AKNKIVNSRK VD UniProtKB: Spindle pole body component 110 |
-Macromolecule #5: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 4 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: Whatman #1 Filter papers used.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 3024 / Average exposure time: 12.0 sec. / Average electron dose: 72.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 47214 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number classes used: 4 Applied symmetry - Helical parameters - Δz: 0.1 Å Applied symmetry - Helical parameters - Δ&Phi: 0 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM (ver. 1.0 Beta) Details: Final reconstruction is on symmetry expanded particles. Number images used: 148911 |
|---|---|
| Segment selection | Number selected: 175500 / Software - Name: EMAN2 / Software - details: e2helixboxer.py Details: Boxed approximately every 3 asymmetric units using a rise of 21 Angstroms, with a box size of 600 pixels (635.4A). |
| Startup model | Type of model: EMDB MAP |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: cisTEM (ver. 1.0 Beta) / Details: Projection matching in cisTEM |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 60 |
|---|---|
| Output model |  PDB-7m2w: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)