+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23607 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the glutamate receptor-like channel AtGLR3.4 | |||||||||||||||
 Map data Map data | Subtracted map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Arabidopsis thaliana / Ion-Channel / glutamate receptor-like channel (GLR) / TRANSPORT PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to acetate / chloroplast membrane / glutamate receptor activity / plastid / ligand-gated monoatomic ion channel activity / cellular response to cold / chloroplast / cellular response to amino acid stimulus / calcium-mediated signaling / cellular response to mechanical stimulus ...cellular response to acetate / chloroplast membrane / glutamate receptor activity / plastid / ligand-gated monoatomic ion channel activity / cellular response to cold / chloroplast / cellular response to amino acid stimulus / calcium-mediated signaling / cellular response to mechanical stimulus / response to wounding / calcium channel activity / calcium ion transport / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
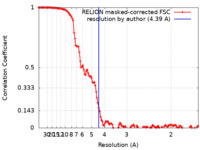
| Method | single particle reconstruction / cryo EM / Resolution: 4.39 Å | |||||||||||||||
 Authors Authors | Gangwar SP / Green MN | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4. Authors: Marriah N Green / Shanti Pal Gangwar / Erwan Michard / Alexander A Simon / Maria Teresa Portes / Juan Barbosa-Caro / Michael M Wudick / Michael A Lizzio / Oleg Klykov / Maria V Yelshanskaya ...Authors: Marriah N Green / Shanti Pal Gangwar / Erwan Michard / Alexander A Simon / Maria Teresa Portes / Juan Barbosa-Caro / Michael M Wudick / Michael A Lizzio / Oleg Klykov / Maria V Yelshanskaya / José A Feijó / Alexander I Sobolevsky /    Abstract: Glutamate receptor-like channels (GLRs) play vital roles in various physiological processes in plants, such as wound response, stomatal aperture control, seed germination, root development, innate ...Glutamate receptor-like channels (GLRs) play vital roles in various physiological processes in plants, such as wound response, stomatal aperture control, seed germination, root development, innate immune response, pollen tube growth, and morphogenesis. Despite the importance of GLRs, knowledge about their molecular organization is limited. Here we use X-ray crystallography and single-particle cryo-EM to solve structures of the Arabidopsis thaliana GLR3.4. Our structures reveal the tetrameric assembly of GLR3.4 subunits into a three-layer domain architecture, reminiscent of animal ionotropic glutamate receptors (iGluRs). However, the non-swapped arrangement between layers of GLR3.4 domains, binding of glutathione through S-glutathionylation of cysteine C205 inside the amino-terminal domain clamshell, unique symmetry, inter-domain interfaces, and ligand specificity distinguish GLR3.4 from representatives of the iGluR family and suggest distinct features of the GLR gating mechanism. Our work elaborates on the principles of GLR architecture and symmetry and provides a molecular template for deciphering GLR-dependent signaling mechanisms in plants. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23607.map.gz emd_23607.map.gz | 10.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23607-v30.xml emd-23607-v30.xml emd-23607.xml emd-23607.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23607_fsc.xml emd_23607_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_23607.png emd_23607.png | 162.6 KB | ||
| Filedesc metadata |  emd-23607.cif.gz emd-23607.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23607 http://ftp.pdbj.org/pub/emdb/structures/EMD-23607 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23607 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23607 | HTTPS FTP |
-Validation report
| Summary document |  emd_23607_validation.pdf.gz emd_23607_validation.pdf.gz | 392.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23607_full_validation.pdf.gz emd_23607_full_validation.pdf.gz | 391.7 KB | Display | |
| Data in XML |  emd_23607_validation.xml.gz emd_23607_validation.xml.gz | 12.8 KB | Display | |
| Data in CIF |  emd_23607_validation.cif.gz emd_23607_validation.cif.gz | 17.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23607 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23607 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23607 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23607 | HTTPS FTP |
-Related structure data
| Related structure data |  7lziMC  7lz0C  7lz1C  7lz2C  7lzhC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23607.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23607.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtracted map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GLR3.4
| Entire | Name: GLR3.4 |
|---|---|
| Components |
|
-Supramolecule #1: GLR3.4
| Supramolecule | Name: GLR3.4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Map displaying ligand binding and trans-membrane domain |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamate receptor 3.4
| Macromolecule | Name: Glutamate receptor 3.4 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 107.317383 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGFLVMIREV SMAKAIRVVL LCVSVLWVVP KECACRSNFS RNSSSSSSSS LRPLRQRPSS VNVGALFTYD SFIGRAAKPA VKAAMDDVN ADQSVLKGIK LNIIFQDSNC SGFIGTMGAL QLMENKVVAA IGPQSSGIAH MISYVANELH VPLLSFGATD P TLSSLQFP ...String: MGFLVMIREV SMAKAIRVVL LCVSVLWVVP KECACRSNFS RNSSSSSSSS LRPLRQRPSS VNVGALFTYD SFIGRAAKPA VKAAMDDVN ADQSVLKGIK LNIIFQDSNC SGFIGTMGAL QLMENKVVAA IGPQSSGIAH MISYVANELH VPLLSFGATD P TLSSLQFP YFLRTTQNDY FQMHAIADFL SYSGWRQVIA IFVDDECGRN GISVLGDVLA KKRSRISYKA AITPGADSSS IR DLLVSVN LMESRVFVVH VNPDSGLNVF SVAKSLGMMA SGYVWIATDW LPTAMDSMEH VDSDTMDLLQ GVVAFRHYTI ESS VKRQFM ARWKNLRPND GFNSYAMYAY DSVWLVARAL DVFFRENNNI TFSNDPNLHK TNGSTIQLSA LSVFNEGEKF MKII LGMNH TGVTGPIQFD SDRNRVNPAY EVLNLEGTAP RTVGYWSNHS GLSVVHPETL YSRPPNTSTA NQRLKGIIYP GEVTK PPRG WVFPNNGKPL RIGVPNRVSY TDYVSKDKNP PGVRGYCIDV FEAAIELLPY PVPRTYILYG DGKRNPSYDN LVNEVV ADN FDVAVGDITI VTNRTRYVDF TQPFIESGLV VVAPVKEAKS SPWSFLKPFT IEMWAVTGGF FLFVGAMVWI LEHRFNQ EF RGPPRRQLIT IFWFSFSTMF FSHRENTVSS LGRFVLIIWL FVVLIINSSY TASLTSILTI RQLTSRIEGI DSLVTSNE P IGVQDGTFAR NYLINELNIL PSRIVPLKDE EQYLSALQRG PNAGGVAAIV DELPYIEVLL TNSNCKFRTV GQEFTRTGW GFAFQRDSPL AVDMSTAILQ LSEEGELEKI HRKWLNYKHE CSMQISNSED SQLSLKSFWG LFLICGITCF MALTVFFWRV FWQYQRLLP ESADEERAGE VSEPSRSGRG SRAPSFKELI KVVDKREAEI KEILKQKSSK KLKSTQSAAG TSQSQHGEIT UniProtKB: Glutamate receptor 3.4 |
-Macromolecule #3: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 3 / Number of copies: 4 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 1mM L-Glutamate was added to the purified protein and incubated on ice for 30 min before specimen preparation.. | ||||||||||||
| Details | Protein extracted and reconstituted in a detergent micelle |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Average exposure time: 2.5 sec. / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)