+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23530 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Marseillevirus heterotrimeric (hexameric) nucleosome | |||||||||
 Map data Map data | Structure of the Marseillevirus Hexamer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Structural Protein/DNA / STRUCTURAL PROTEIN / STRUCTURAL PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of chromatin / protein heterodimerization activity / DNA binding Similarity search - Function | |||||||||
| Biological species | synthetic construct (others) /  Marseillevirus marseillevirus Marseillevirus marseillevirus | |||||||||
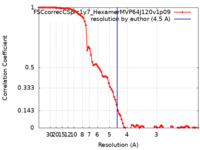
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Valencia-Sanchez MI / Abini-Agbomson S | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: The structure of a virus-encoded nucleosome. Authors: Marco Igor Valencia-Sánchez / Stephen Abini-Agbomson / Miao Wang / Rachel Lee / Nikita Vasilyev / Jenny Zhang / Pablo De Ioannes / Bernard La Scola / Paul Talbert / Steve Henikoff / Evgeny ...Authors: Marco Igor Valencia-Sánchez / Stephen Abini-Agbomson / Miao Wang / Rachel Lee / Nikita Vasilyev / Jenny Zhang / Pablo De Ioannes / Bernard La Scola / Paul Talbert / Steve Henikoff / Evgeny Nudler / Albert Erives / Karim-Jean Armache /   Abstract: Certain large DNA viruses, including those in the Marseilleviridae family, encode histones. Here we show that fused histone pairs Hβ-Hα and Hδ-Hγ from Marseillevirus are structurally analogous to ...Certain large DNA viruses, including those in the Marseilleviridae family, encode histones. Here we show that fused histone pairs Hβ-Hα and Hδ-Hγ from Marseillevirus are structurally analogous to the eukaryotic histone pairs H2B-H2A and H4-H3. These viral histones form 'forced' heterodimers, and a heterotetramer of four such heterodimers assembles DNA to form structures virtually identical to canonical eukaryotic nucleosomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23530.map.gz emd_23530.map.gz | 22.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23530-v30.xml emd-23530-v30.xml emd-23530.xml emd-23530.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23530_fsc.xml emd_23530_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_23530.png emd_23530.png | 198.7 KB | ||
| Filedesc metadata |  emd-23530.cif.gz emd-23530.cif.gz | 6.4 KB | ||
| Others |  emd_23530_half_map_1.map.gz emd_23530_half_map_1.map.gz emd_23530_half_map_2.map.gz emd_23530_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23530 http://ftp.pdbj.org/pub/emdb/structures/EMD-23530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23530 | HTTPS FTP |
-Related structure data
| Related structure data |  7lv9MC  7lv8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23530.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23530.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the Marseillevirus Hexamer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
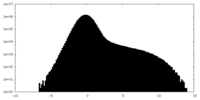
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map A Structure of the Marseillevirus Hexamer
| File | emd_23530_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A Structure of the Marseillevirus Hexamer | ||||||||||||
| Projections & Slices |
| ||||||||||||
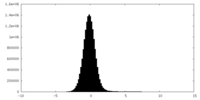
| Density Histograms |
-Half map: Half map B Structure of the Marseillevirus Hexamer
| File | emd_23530_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B Structure of the Marseillevirus Hexamer | ||||||||||||
| Projections & Slices |
| ||||||||||||
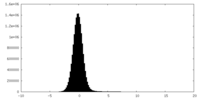
| Density Histograms |
- Sample components
Sample components
-Entire : Marseillevirus heterotrimeric (hexameric) nucleosome
| Entire | Name: Marseillevirus heterotrimeric (hexameric) nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: Marseillevirus heterotrimeric (hexameric) nucleosome
| Supramolecule | Name: Marseillevirus heterotrimeric (hexameric) nucleosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: virus-encoded histone doublets Marseillevirus |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Histone doublet Delta-Gamma (Delta)
| Macromolecule | Name: Histone doublet Delta-Gamma (Delta) / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Marseillevirus marseillevirus Marseillevirus marseillevirus |
| Molecular weight | Theoretical: 10.462416 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LADHVSVGET QIPKASTQHL LRKAGSLSAA GDTEVPIRGF VHMKLHKLVQ KSLLAMQLAK RKTIMKSDVK KAAELMHLPV FAIPTKDSG AKGSVFLS UniProtKB: Histone H3 |
-Macromolecule #2: Histone doublet Delta-Gamma (Gamma)
| Macromolecule | Name: Histone doublet Delta-Gamma (Gamma) / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Marseillevirus marseillevirus Marseillevirus marseillevirus |
| Molecular weight | Theoretical: 11.7676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CRQKGAGSAG TGSETNSQEV RSQMRSTCLI IPKERFRTMA KEISKKEGHD VHIAEAALDM LQVIVESCTV RLLEKALVIT YSGKRTRVT SKDIETAFML EHGPLLE UniProtKB: Histone H3 |
-Macromolecule #3: Histone doublet Beta-Alpha (Beta)
| Macromolecule | Name: Histone doublet Beta-Alpha (Beta) / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Marseillevirus marseillevirus Marseillevirus marseillevirus |
| Molecular weight | Theoretical: 11.228021 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATQKETTRK RDKSVNFRLG LRNMLAQIHP DISVQTEALS ELSNIAVFLG KKISHGAVTL LPEGTKTIKS SAVLLAAGDL YGKDLGRHA VGEMTKAVTR YGSAK UniProtKB: Histone H2B/H2A fusion protein |
-Macromolecule #4: Histone doublet Beta-Alpha (Alpha)
| Macromolecule | Name: Histone doublet Beta-Alpha (Alpha) / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Marseillevirus marseillevirus Marseillevirus marseillevirus |
| Molecular weight | Theoretical: 17.675625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ESKEGSRSSK AKLQISVARS ERLLREHGGC SRVSEGAAVA LAAAIEYFMG EVLELAGNAA RDSKKVRISV KHITLAIQND AALFAVVGK GVFSGAGVSL ISVPIPRKKA RKTTEKEASS PKKKAAPKKK KAASKQKKSL SDKELAKLTK KELAKYEKEQ G MSPGY UniProtKB: Histone H2B/H2A fusion protein |
-Macromolecule #5: DNA (96-MER)
| Macromolecule | Name: DNA (96-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 29.08757 KDa |
| Sequence | String: (DG)(DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA)(DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DG)(DA)(DT)(DT)(DC)(DT)(DC) (DC) (DC)(DC)(DC)(DG)(DC)(DG) ...String: (DG)(DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA)(DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DG)(DA)(DT)(DT)(DC)(DT)(DC) (DC) (DC)(DC)(DC)(DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DA) (DA)(DG) (DG)(DG)(DG)(DA)(DT)(DT)(DA) (DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT)(DC) (DT)(DC)(DC) (DA)(DG)(DG)(DC)(DA)(DC) (DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT) |
-Macromolecule #6: DNA (96-MER)
| Macromolecule | Name: DNA (96-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 29.527826 KDa |
| Sequence | String: (DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA)(DC)(DG) (DT)(DG)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DC)(DT)(DA)(DG)(DG)(DG)(DA)(DG)(DT) (DA)(DA)(DT)(DC)(DC)(DC)(DC)(DT)(DT)(DG) (DG) (DC)(DG)(DG)(DT)(DT)(DA) ...String: (DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA)(DC)(DG) (DT)(DG)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DC)(DT)(DA)(DG)(DG)(DG)(DA)(DG)(DT) (DA)(DA)(DT)(DC)(DC)(DC)(DC)(DT)(DT)(DG) (DG) (DC)(DG)(DG)(DT)(DT)(DA)(DA)(DA) (DA)(DC)(DG)(DC)(DG)(DG)(DG)(DG)(DG)(DA) (DG)(DA) (DA)(DT)(DC)(DC)(DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG)(DT)(DT)(DT)(DA) (DA)(DG)(DC) (DG)(DG)(DT)(DG)(DC)(DT) (DA)(DG)(DA)(DG)(DC)(DT)(DG)(DT)(DC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 4503 / Average exposure time: 2.5 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7lv9: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)