[English] 日本語
 Yorodumi
Yorodumi- EMDB-23521: Prefusion RSV F glycoprotein bound by neutralizing site V-directe... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23521 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
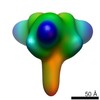
| Title | Prefusion RSV F glycoprotein bound by neutralizing site V-directed antibody ADI-14442 | |||||||||
 Map data Map data | Trimeric prefusion RSV F protein bound by three copies of Fab ADI-14442 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | fusion / immunoglobulin / public clonotype / convergent recognition / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated induction of syncytium formation / host cell Golgi membrane / entry receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Respiratory syncytial virus A2 / Respiratory syncytial virus A2 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Gilman MSA / McLellan JS | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2021 Journal: Immunity / Year: 2021Title: Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses. Authors: Maryam Mukhamedova / Daniel Wrapp / Chen-Hsiang Shen / Morgan S A Gilman / Tracy J Ruckwardt / Chaim A Schramm / Larissa Ault / Lauren Chang / Alexandrine Derrien-Colemyn / Sarah A M Lucas / ...Authors: Maryam Mukhamedova / Daniel Wrapp / Chen-Hsiang Shen / Morgan S A Gilman / Tracy J Ruckwardt / Chaim A Schramm / Larissa Ault / Lauren Chang / Alexandrine Derrien-Colemyn / Sarah A M Lucas / Amy Ransier / Samuel Darko / Emily Phung / Lingshu Wang / Yi Zhang / Scott A Rush / Bharat Madan / Guillaume B E Stewart-Jones / Pamela J Costner / LaSonji A Holman / Somia P Hickman / Nina M Berkowitz / Nicole A Doria-Rose / Kaitlyn M Morabito / Brandon J DeKosky / Martin R Gaudinski / Grace L Chen / Michelle C Crank / John Misasi / Nancy J Sullivan / Daniel C Douek / Peter D Kwong / Barney S Graham / Jason S McLellan / John R Mascola /  Abstract: An effective vaccine for respiratory syncytial virus (RSV) is an unrealized public health goal. A single dose of the prefusion-stabilized fusion (F) glycoprotein subunit vaccine (DS-Cav1) ...An effective vaccine for respiratory syncytial virus (RSV) is an unrealized public health goal. A single dose of the prefusion-stabilized fusion (F) glycoprotein subunit vaccine (DS-Cav1) substantially increases serum-neutralizing activity in healthy adults. We sought to determine whether DS-Cav1 vaccination induces a repertoire mirroring the pre-existing diversity from natural infection or whether antibody lineages targeting specific epitopes predominate. We evaluated RSV F-specific B cell responses before and after vaccination in six participants using complementary B cell sequencing methodologies and identified 555 clonal lineages. DS-Cav1-induced lineages recognized the prefusion conformation of F (pre-F) and were genetically diverse. Expressed antibodies recognized all six antigenic sites on the pre-F trimer. We identified 34 public clonotypes, and structural analysis of two antibodies from a predominant clonotype revealed a common mode of recognition. Thus, vaccination with DS-Cav1 generates a diverse polyclonal response targeting the antigenic sites on pre-F, supporting the development and advanced testing of pre-F-based vaccines against RSV. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23521.map.gz emd_23521.map.gz | 8.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23521-v30.xml emd-23521-v30.xml emd-23521.xml emd-23521.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23521_fsc.xml emd_23521_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23521.png emd_23521.png | 44.3 KB | ||
| Filedesc metadata |  emd-23521.cif.gz emd-23521.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23521 http://ftp.pdbj.org/pub/emdb/structures/EMD-23521 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23521 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23521 | HTTPS FTP |
-Related structure data
| Related structure data |  7lueMC  7lucC  7ludC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23521.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23521.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Trimeric prefusion RSV F protein bound by three copies of Fab ADI-14442 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.075 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Trimeric prefusion RSV F glycoprotein bound by three molecules of...
| Entire | Name: Trimeric prefusion RSV F glycoprotein bound by three molecules of Fab ADI-14442. |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric prefusion RSV F glycoprotein bound by three molecules of...
| Supramolecule | Name: Trimeric prefusion RSV F glycoprotein bound by three molecules of Fab ADI-14442. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Single molecule of trimeric respiratory syncytial virus fusion protein ectodomain (RSV F) stabilized in the prefusion conformation and fused to the T4 fibritin (foldon) trimerization motif ...Details: Single molecule of trimeric respiratory syncytial virus fusion protein ectodomain (RSV F) stabilized in the prefusion conformation and fused to the T4 fibritin (foldon) trimerization motif is bound by three molecules of Fab ADI-14442. |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
-Supramolecule #2: Trimeric prefusion RSV F glycoprotein
| Supramolecule | Name: Trimeric prefusion RSV F glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: Single molecule of trimeric respiratory syncytial virus fusion protein ectodomain (RSV F) stabilized in the prefusion conformation and fused to the T4 fibritin (foldon) trimerization motif. |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
-Supramolecule #3: Fab ADI-14442
| Supramolecule | Name: Fab ADI-14442 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 Details: Three molecules of Fab ADI-14442, each composed of a heavy and a light chain, are bound to a single molecule of trimeric respiratory syncytial virus fusion protein ectodomain (RSV F) ...Details: Three molecules of Fab ADI-14442, each composed of a heavy and a light chain, are bound to a single molecule of trimeric respiratory syncytial virus fusion protein ectodomain (RSV F) stabilized in the prefusion conformation and fused to the T4 fibritin (foldon) trimerization motif. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fusion glycoprotein F0
| Macromolecule | Name: Fusion glycoprotein F0 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 61.132672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QNITEEFYQS TCSAVSKGYL SALRTGWYTS VITIELSNIK EIKCNGTDAK VKLIKQELDK YKNAVTELQL LMQSTPATNN RARRELPRF MNYTLNNAKK TNVTLSKKRK RRFLGFLLGV GSAIASGVAV SKVLHLEGEV NKIKSALLST NKAVVSLSNG V SVLTSKVL ...String: QNITEEFYQS TCSAVSKGYL SALRTGWYTS VITIELSNIK EIKCNGTDAK VKLIKQELDK YKNAVTELQL LMQSTPATNN RARRELPRF MNYTLNNAKK TNVTLSKKRK RRFLGFLLGV GSAIASGVAV SKVLHLEGEV NKIKSALLST NKAVVSLSNG V SVLTSKVL DLKNYIDKQL LPIVNKQSCS IPNIETVIEF QQKNNRLLEI TREFSVNAGV TTPVSTYMLT NSELLSLIND MP ITNDQKK LMSNNVQIVR QQSYSIMSII KEEVLAYVVQ LPLYGVIDTP CWKLHTSPLC TTNTKEGSNI CLTRTDRGWY CDN AGSVSF FPQAETCKVQ SNRVFCDTMN SLTLPSEVNL CNVDIFNPKY DCKIMTSKTD VSSSVITSLG AIVSCYGKTK CTAS NKNRG IIKTFSNGCD YVSNKGVDTV SVGNTLYYVN KQEGKSLYVK GEPIINFYDP LVFPSDEFDA SISQVNEKIN QSLAF IRKS DELLSAIGGY IPEAPRDGQA YVRKDGEWVL LSTFLGSLEV LFQGPGHHHH HHHHSAWSHP QFEK UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #2: Heavy chain of human antibody Fab ADI-14442
| Macromolecule | Name: Heavy chain of human antibody Fab ADI-14442 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.805615 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGSE VKKPGASVKV SCKASGYRFS NYGISWVRQA PGQGLEWMGW ISAYNGNIKY GNNLQGRVTV TTDTSTATAY MEVRSLTSD DTAVYYCARD VPADGVHFMD VWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT ...String: QVQLVQSGSE VKKPGASVKV SCKASGYRFS NYGISWVRQA PGQGLEWMGW ISAYNGNIKY GNNLQGRVTV TTDTSTATAY MEVRSLTSD DTAVYYCARD VPADGVHFMD VWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCD |
-Macromolecule #3: Light chain of human antibody Fab ADI-14442
| Macromolecule | Name: Light chain of human antibody Fab ADI-14442 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.100822 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVVMTQSPLS LPVTLGQPAS ISCRSSQSLV HSDTNTYLNW FQQRPGQSPR RLIYKVSNRD SGVPDRFSGS GSGTTFTLKI SRVEAEDVG IYYCMQGSHW APTFGQGTKV EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: DVVMTQSPLS LPVTLGQPAS ISCRSSQSLV HSDTNTYLNW FQQRPGQSPR RLIYKVSNRD SGVPDRFSGS GSGTTFTLKI SRVEAEDVG IYYCMQGSHW APTFGQGTKV EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 9.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 22500 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















