+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23496 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of the SLFN12-PDE3A complex: SLFN12 body refinement | |||||||||
 Map data Map data | Body output from Relion Multibody refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / velcrin / molecular glue / DNMDP / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationrRNA catabolic process / RNA nuclease activity / apoptotic signaling pathway / ribosome binding / Hydrolases; Acting on ester bonds / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
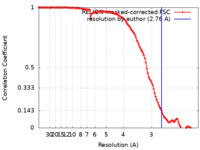
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||
 Authors Authors | Fuller JR / Garvie CW / Lemke CT | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase. Authors: Colin W Garvie / Xiaoyun Wu / Malvina Papanastasiou / Sooncheol Lee / James Fuller / Gavin R Schnitzler / Steven W Horner / Andrew Baker / Terry Zhang / James P Mullahoo / Lindsay Westlake / ...Authors: Colin W Garvie / Xiaoyun Wu / Malvina Papanastasiou / Sooncheol Lee / James Fuller / Gavin R Schnitzler / Steven W Horner / Andrew Baker / Terry Zhang / James P Mullahoo / Lindsay Westlake / Stephanie H Hoyt / Marcus Toetzl / Matthew J Ranaghan / Luc de Waal / Joseph McGaunn / Bethany Kaplan / Federica Piccioni / Xiaoping Yang / Martin Lange / Adrian Tersteegen / Donald Raymond / Timothy A Lewis / Steven A Carr / Andrew D Cherniack / Christopher T Lemke / Matthew Meyerson / Heidi Greulich /   Abstract: DNMDP and related compounds, or velcrins, induce complex formation between the phosphodiesterase PDE3A and the SLFN12 protein, leading to a cytotoxic response in cancer cells that express elevated ...DNMDP and related compounds, or velcrins, induce complex formation between the phosphodiesterase PDE3A and the SLFN12 protein, leading to a cytotoxic response in cancer cells that express elevated levels of both proteins. The mechanisms by which velcrins induce complex formation, and how the PDE3A-SLFN12 complex causes cancer cell death, are not fully understood. Here, we show that PDE3A and SLFN12 form a heterotetramer stabilized by binding of DNMDP. Interactions between the C-terminal alpha helix of SLFN12 and residues near the active site of PDE3A are required for complex formation, and are further stabilized by interactions between SLFN12 and DNMDP. Moreover, we demonstrate that SLFN12 is an RNase, that PDE3A binding increases SLFN12 RNase activity, and that SLFN12 RNase activity is required for DNMDP response. This new mechanistic understanding will facilitate development of velcrin compounds into new cancer therapies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23496.map.gz emd_23496.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23496-v30.xml emd-23496-v30.xml emd-23496.xml emd-23496.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23496_fsc.xml emd_23496_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_23496.png emd_23496.png | 92.6 KB | ||
| Masks |  emd_23496_msk_1.map emd_23496_msk_1.map | 129.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23496.cif.gz emd-23496.cif.gz | 7.2 KB | ||
| Others |  emd_23496_additional_1.map.gz emd_23496_additional_1.map.gz emd_23496_half_map_1.map.gz emd_23496_half_map_1.map.gz emd_23496_half_map_2.map.gz emd_23496_half_map_2.map.gz | 75.9 MB 97.3 MB 97.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23496 http://ftp.pdbj.org/pub/emdb/structures/EMD-23496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23496 | HTTPS FTP |
-Related structure data
| Related structure data |  7lreMC  7kweC  7l27C  7l28C  7l29C  7lrcC  7lrdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23496.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23496.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body output from Relion Multibody refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23496_msk_1.map emd_23496_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
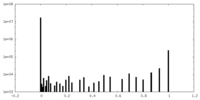
| Density Histograms |
-Additional map: Sharpened by an automatically-determined B-factor (-100.32) and filtered...
| File | emd_23496_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened by an automatically-determined B-factor (-100.32) and filtered to local resolution, using the Relion local resolution implementation. Model was primarily refined against this map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
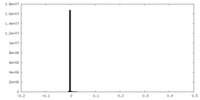
| Density Histograms |
-Half map: Body half-map 1 from Relion Multibody refinement
| File | emd_23496_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body half-map 1 from Relion Multibody refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
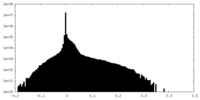
| Density Histograms |
-Half map: Body half-map 2 from Relion Multibody refinement
| File | emd_23496_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body half-map 2 from Relion Multibody refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
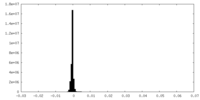
| Density Histograms |
- Sample components
Sample components
-Entire : SLFN12-PDE3A complex, SLFN12 body
| Entire | Name: SLFN12-PDE3A complex, SLFN12 body |
|---|---|
| Components |
|
-Supramolecule #1: SLFN12-PDE3A complex, SLFN12 body
| Supramolecule | Name: SLFN12-PDE3A complex, SLFN12 body / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Schlafen family member 12
| Macromolecule | Name: Schlafen family member 12 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.383734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGGGSMNISV DLETNYAELV LDVGRVTLGE NSRKKMKDCK LRKKQNESVS RAMCALLNSG GGVIKAEIEN EDYSYTKDGI GLDLENSFS NILLFVPEYL DFMQNGNYFL IFVKSWSLNT SGLRITTLSS NLYKRDITSA KVMNATAALE FLKDMKKTRG R LYLRPELL ...String: GGGGSMNISV DLETNYAELV LDVGRVTLGE NSRKKMKDCK LRKKQNESVS RAMCALLNSG GGVIKAEIEN EDYSYTKDGI GLDLENSFS NILLFVPEYL DFMQNGNYFL IFVKSWSLNT SGLRITTLSS NLYKRDITSA KVMNATAALE FLKDMKKTRG R LYLRPELL AKRPCVDIQE ENNMKALAGV FFDRTELDRK EKLTFTESTH VEIKNFSTEK LLQRIKEILP QYVSAFANTD GG YLFIGLN EDKEIIGFKA EMSDLDDLER EIEKSIRKMP VHHFCMEKKK INYSCKFLGV YDKGSLCGYV CALRVERFCC AVF AKEPDS WHVKDNRVMQ LTRKEWIQFM VEAEPKFSSS YEEVISQINT SLPAPHSWPL LEWQRQRHHC PGLSGRITYT PENL CRKLF LQHEGLKQLI CEEMDSVRKG SLIFSRSWSV DLGLQENHKV LCDALLISQD SPPVLYTFHM VQDEEFKGYS TQTAL TLKQ KLAKIGGYTK KVCVMTKIFY LSPEGMTSCQ YDLRSQVIYP ESYYFTRRKY LLKALFKALK RLKSLRDQFS FAENLY QII GIDCFQKNDK KMFKSCRRLT UniProtKB: Ribonuclease SLFN12 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 0.0038% NP-40s detergent (CAS 9016-45-9) added immediately prior to plunge freezing | |||||||||||||||
| Grid | Model: C-flat / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: 4.5 second blot time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 3163 / Average electron dose: 50.0 e/Å2 Details: Exposures were collected in super-resolution mode, as movies fractionated over 40 frames |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The atomic model (including per-residue ADP/B-factors) was refined in Phenix against the sharpened/local resolution-filtered map |
| Refinement | Space: REAL |
| Output model |  PDB-7lre: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)