+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23408 | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chicken Scap D435V L1-L7 domain / Fab complex focused map | ||||||||||||||||||||||||||||||||||||
 Map data Map data | Map of L1L7/Fab domain from focused classification | ||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Cholesterol / LIPID BINDING PROTEIN | ||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationRegulation of cholesterol biosynthesis by SREBP (SREBF) / SREBP-SCAP complex / regulation of cholesterol biosynthetic process / sterol binding / SREBP signaling pathway / regulation of fatty acid biosynthetic process / cholesterol metabolic process / ER to Golgi transport vesicle membrane / positive regulation of cholesterol biosynthetic process / response to insulin ...Regulation of cholesterol biosynthesis by SREBP (SREBF) / SREBP-SCAP complex / regulation of cholesterol biosynthetic process / sterol binding / SREBP signaling pathway / regulation of fatty acid biosynthetic process / cholesterol metabolic process / ER to Golgi transport vesicle membrane / positive regulation of cholesterol biosynthetic process / response to insulin / response to hypoxia / immune response / Golgi membrane / endoplasmic reticulum membrane Similarity search - Function | ||||||||||||||||||||||||||||||||||||
| Biological species |   | ||||||||||||||||||||||||||||||||||||
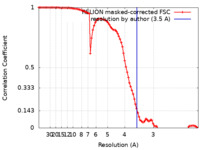
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||||||||||||||||||||||||||
 Authors Authors | Kober DL / Radhakrishnan A | ||||||||||||||||||||||||||||||||||||
| Funding support |  United States, United States,  France, 11 items France, 11 items
| ||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing. Authors: Daniel L Kober / Arun Radhakrishnan / Joseph L Goldstein / Michael S Brown / Lindsay D Clark / Xiao-Chen Bai / Daniel M Rosenbaum /  Abstract: The cholesterol-sensing protein Scap induces cholesterol synthesis by transporting membrane-bound transcription factors called sterol regulatory element-binding proteins (SREBPs) from the endoplasmic ...The cholesterol-sensing protein Scap induces cholesterol synthesis by transporting membrane-bound transcription factors called sterol regulatory element-binding proteins (SREBPs) from the endoplasmic reticulum (ER) to the Golgi apparatus for proteolytic activation. Transport requires interaction between Scap's two ER luminal loops (L1 and L7), which flank an intramembrane sterol-sensing domain (SSD). Cholesterol inhibits Scap transport by binding to L1, which triggers Scap's binding to Insig, an ER retention protein. Here we used cryoelectron microscopy (cryo-EM) to elucidate two structures of full-length chicken Scap: (1) a wild-type free of Insigs and (2) mutant Scap bound to chicken Insig without cholesterol. Strikingly, L1 and L7 intertwine tightly to form a globular domain that acts as a luminal platform connecting the SSD to the rest of Scap. In the presence of Insig, this platform undergoes a large rotation accompanied by rearrangement of Scap's transmembrane helices. We postulate that this conformational change halts Scap transport of SREBPs and inhibits cholesterol synthesis. | ||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23408.map.gz emd_23408.map.gz | 70.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23408-v30.xml emd-23408-v30.xml emd-23408.xml emd-23408.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23408_fsc.xml emd_23408_fsc.xml | 9.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_23408.png emd_23408.png | 29.4 KB | ||
| Filedesc metadata |  emd-23408.cif.gz emd-23408.cif.gz | 7.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23408 http://ftp.pdbj.org/pub/emdb/structures/EMD-23408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23408 | HTTPS FTP |
-Related structure data
| Related structure data |  7lkhMC  7lkfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23408.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23408.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of L1L7/Fab domain from focused classification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ternary complex of Scap with Fab fragment
| Entire | Name: Ternary complex of Scap with Fab fragment |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Scap with Fab fragment
| Supramolecule | Name: Ternary complex of Scap with Fab fragment / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Fab fragment generated by proteolytic cleavage of IgG antibody |
|---|---|
| Molecular weight | Theoretical: 172 KDa |
-Supramolecule #2: Scap L1-L7 domain
| Supramolecule | Name: Scap L1-L7 domain / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50 KDa |
-Supramolecule #3: 4G10 Fab
| Supramolecule | Name: 4G10 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 Details: Fab fragment generated by proteolytic cleavage of IgG antibody |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Sterol regulatory element-binding protein cleavage-activating protein
| Macromolecule | Name: Sterol regulatory element-binding protein cleavage-activating protein type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 148.364828 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PAMTLTEKLR ERISRAFYNH GLLCASYPIP IILFTGLCIL ACCYPLLKLP LPGTGPVEFS TPVKDYFPPS PDVVSQQGDL SERPDWYVG APVAYIQQIF VKATVSPWQK NFLAVDVFRS PLSRVFQLVE EIRNHALRDS SGVKSLEEVC LQVTDLLPGL K KLRNLLPE ...String: PAMTLTEKLR ERISRAFYNH GLLCASYPIP IILFTGLCIL ACCYPLLKLP LPGTGPVEFS TPVKDYFPPS PDVVSQQGDL SERPDWYVG APVAYIQQIF VKATVSPWQK NFLAVDVFRS PLSRVFQLVE EIRNHALRDS SGVKSLEEVC LQVTDLLPGL K KLRNLLPE HGCLLLSPGN FWQNDRERFN ADPDIIKTIH QHEPKALQTS ATLKDLLFGL PGKYSGVNLY NRKRVVSYTV TL GLQRYDS RFLSSLRSRL KLLHPSPNCT LREDSIVHVH FKEEIGIAEL IPLVTTYIIL FAYIYFSTRK IDMVKSKWGL ALA AVVTVL SSLLMSVGLC TLFGLTPTLN GGEIFPYLVV VIGLENVLVL TKSVVSTPVD LEVKLRIAQG LSNESWSIMK NMAT ELGII LIGYFTLVPA IQEFCLFAVV GLVSVFFLQM LFFTTVLSID IRRMELADLN KRLPAEACLP PAKPASRSQR YERQP AVRP ATPHTITLQP SSFRNLRLPK RLRVIYFFAR TRLAQRLIMA GTVIWIGILV YTDPAGLRTY LTSQVTEQSP LGEAGL PPM PVPGGVLPAG DPKIDLSVFP SDPIQLSENQ TQQREQQAGL EPLGRLETNQ HSWAQGPEGR GNGQTELGTE AEVTWGA ED EEIWRKLSFR HWPSLFSYYN ITLAKRYISI LPAIPVTLYL NPQEALEVRH PQEANRYHPF LSSSGGKLNA EAQPDQTS S RLQGHRDVTL YKVAALGLAS GILLVLLLFC LYRLLCPKNY GQNGLSHSRR RRGDLPCDDY GYSPPETEIV PLVLRGHLM DIECLASDGM LLVSCCLVGQ IRVWDAQTGD CLTVIPKPRL RRDSSGIFDY QESWDHSPDG KTGLDDSFES SHQLKRMLSP PQPPLFCDQ PDLTSLIDTN FSEQVKVAES EPRLRAVGGR QKEAGYDFSS LVGKVYEEHS TSNCMNFGGL SAPHGQAGFC V GGSTARSL GCGSEEGGCG GRRRSLGDES LSGFDKSSPL PSWGGDFESS VWSLDLQGNL IVAGRSNGKL EVWDAIEGTL RS SNDESQS GITALVFLNN RIVAARLNGS LDFFSLETHT SLNHLQFRGA PSRSSIPSSP LFSSSDVIVC QLTHTVSCAH QKP ITALKA AAGRLVTGSQ DHTLRVFRLE DSCCLFTLQG HSGAITAVYI DQTMVLASGG QDGAICLWDV LTGSKVSHMY AHRG DVTSL TCTTSCVISS GLDDVISIWD RSSGIKLYSI QQEMGCGSSL GVISDNLLVT GGQGCVSFWD IGYGDLLQTV YLGKS NESQ PARQILVLEN AAIVCNFGSE LSLVYVPSVL EKLDDYKDDD DKGSDYKDDD DKGSDYKDDD DK UniProtKB: Sterol regulatory element-binding protein cleavage-activating protein |
-Macromolecule #2: 4G10 Fab heavy chain
| Macromolecule | Name: 4G10 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.94266 KDa |
| Sequence | String: TGVHSEVQLQ QSGAELVRPG ASVKLSCTAS GFKIKDDYIH WVKQRPEQGL EWIGRIDPAN GHTRYAPKFQ DKATITADTS SNTAYLQLS SLTSEDTAVY YCTRYNDYDA FYFDYWGQGT TLTVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS ...String: TGVHSEVQLQ QSGAELVRPG ASVKLSCTAS GFKIKDDYIH WVKQRPEQGL EWIGRIDPAN GHTRYAPKFQ DKATITADTS SNTAYLQLS SLTSEDTAVY YCTRYNDYDA FYFDYWGQGT TLTVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPKSC DKT |
-Macromolecule #3: 4G10 Fab kappa chain
| Macromolecule | Name: 4G10 Fab kappa chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.667178 KDa |
| Sequence | String: DIQMTQTTSS LSASLGDRVT ISCRASQDIR NYLNWYQQKP DGTVKLLIYY TSRLHSGVPS RFSGSGSGTD YSLTISNLEQ EDIATYFCQ QTNTLPWTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQTTSS LSASLGDRVT ISCRASQDIR NYLNWYQQKP DGTVKLLIYY TSRLHSGVPS RFSGSGSGTD YSLTISNLEQ EDIATYFCQ QTNTLPWTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)