[English] 日本語
 Yorodumi
Yorodumi- EMDB-23339: Cryo-EM map of E. coli P pilus tip assembly intermediate PapC-Pap... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23339 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapG in the first conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / Uropathogenic Escherichia coli / chaperone-usher / P pilus / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcell adhesion involved in single-species biofilm formation / pilus / : / cell wall organization / outer membrane-bounded periplasmic space / carbohydrate binding / cell adhesion / extracellular region Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Du M / Yuan Z | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis. Authors: Minge Du / Zuanning Yuan / Glenn T Werneburg / Nadine S Henderson / Hemil Chauhan / Amanda Kovach / Gongpu Zhao / Jessica Johl / Huilin Li / David G Thanassi /  Abstract: Uropathogenic Escherichia coli assemble surface structures termed pili or fimbriae to initiate infection of the urinary tract. P pili facilitate bacterial colonization of the kidney and ...Uropathogenic Escherichia coli assemble surface structures termed pili or fimbriae to initiate infection of the urinary tract. P pili facilitate bacterial colonization of the kidney and pyelonephritis. P pili are assembled through the conserved chaperone-usher pathway. Much of the structural and functional understanding of the chaperone-usher pathway has been gained through investigations of type 1 pili, which promote binding to the bladder and cystitis. In contrast, the structural basis for P pilus biogenesis at the usher has remained elusive. This is in part due to the flexible and variable-length P pilus tip fiber, creating structural heterogeneity, and difficulties isolating stable P pilus assembly intermediates. Here, we circumvent these hindrances and determine cryo-electron microscopy structures of the activated PapC usher in the process of secreting two- and three-subunit P pilus assembly intermediates, revealing processive steps in P pilus biogenesis and capturing new conformational dynamics of the usher assembly machine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23339.map.gz emd_23339.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23339-v30.xml emd-23339-v30.xml emd-23339.xml emd-23339.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23339.png emd_23339.png | 128 KB | ||
| Filedesc metadata |  emd-23339.cif.gz emd-23339.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23339 http://ftp.pdbj.org/pub/emdb/structures/EMD-23339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23339 | HTTPS FTP |
-Related structure data
| Related structure data |  7lhgMC  7lhhC  7lhiC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23339.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23339.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.029 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
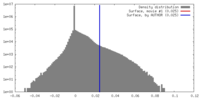
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PapCDKG
| Entire | Name: PapCDKG |
|---|---|
| Components |
|
-Supramolecule #1: PapCDKG
| Supramolecule | Name: PapCDKG / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: P fimbrial usher protein PapC
| Macromolecule | Name: P fimbrial usher protein PapC / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 88.746297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VEFNTDVLDA ADKKNIDFTR FSEAGYVLPG QYLLDVIVNG QSISPASLQI SFVEPQSSGD KAEKKLPQAC LTSDMVRLMG LTAESLDKV VYWHDGQCAD FHGLPGVDIR PDTGAGVLRI NMPQARLEYS DATWLPPSRW DDGIPGLMLD YNLNGTVSRN Y QGGDSHQF ...String: VEFNTDVLDA ADKKNIDFTR FSEAGYVLPG QYLLDVIVNG QSISPASLQI SFVEPQSSGD KAEKKLPQAC LTSDMVRLMG LTAESLDKV VYWHDGQCAD FHGLPGVDIR PDTGAGVLRI NMPQARLEYS DATWLPPSRW DDGIPGLMLD YNLNGTVSRN Y QGGDSHQF SYNGTVGGNL GPWRLRADYQ GSQEQSRYNG EKTTNRNFTW SRFYLFRAIP RWRANLTLGE NNINSDIFRS WS YTGASLE SDDRMLPPRL RGYAPQITGI AETNARVVVS QQGRVLYDSM VPAGPFSIQD LDSSVRGRLD VEVIEQNGRK KTF QVDTAS VPYLTRPGQV RYKLVSGRSR GYGHETEGPV FATGEASWGL SNQWSLYGGA VLAGDYNALA AGAGWDLGVP GTLS ADITQ SVARIEGERT FQGKSWRLSY SKRFDNADAD ITFAGYRFSE RNYMTMEQYL NARYRNDYSS REKEMYTVTL NKNVA DWNT SFNLQYSRQT YWDIRKTDYY TVSVNRYFNV FGLQGVAVGL SASRSKYLGR DNDSAYLRIS VPLGTGTASY SGSMSN DRY VNMAGYTDTF NDGLDSYSLN AGLNSGGGLT SQRQINAYYS HRSPLANLSA NIASLQKGYT SFGVSASGGA TITGKDA AL HAGGMSGGTR LLVDTDGVGG VPVDGGQVVT NRWGTGVVTD ISSYYRNTTS VDLKRLPDDV EATRSVVESA LTEGAIGY R KFSVLKGKRL FAILRLADGS QPPFGASVTS EKGRELGMVA DEGLAWLSGV TPGETLSVNW DGKIQCQVNV PETAISDQQ LLLPCTPQK UniProtKB: UNIPROTKB: A0A773A954 |
-Macromolecule #2: Chaperone protein PapD
| Macromolecule | Name: Chaperone protein PapD / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.833746 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIRKKILMAA IPLFVISGAD AAVSLDRTRA VFDGSEKSMT LDISNDNKQL PYLAQAWIEN ENQEKIITGP VIATPPVQRL EPGAKSMVR LSTTPDISKL PQDRESLFYF NLREIPPRSE KANVLQIALQ TKIKLFYRPA AIKTRPNEVW QDQLILNKVS G GYRIENPT ...String: MIRKKILMAA IPLFVISGAD AAVSLDRTRA VFDGSEKSMT LDISNDNKQL PYLAQAWIEN ENQEKIITGP VIATPPVQRL EPGAKSMVR LSTTPDISKL PQDRESLFYF NLREIPPRSE KANVLQIALQ TKIKLFYRPA AIKTRPNEVW QDQLILNKVS G GYRIENPT PYYVTVIGLG GSEKQAEEGE FETVMLSPRS EQTVKSANYN TPYLSYINDY GGRPVLSFIC NGSRCSVKKE K UniProtKB: Chaperone protein PapD |
-Macromolecule #3: Fimbrial adapter PapK
| Macromolecule | Name: Fimbrial adapter PapK / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.895492 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIKSTGALLL FAALSAGQAI ASDVAFRGNL LDRPCHVSGD SLNKHVVFKT RASRDFWYPP GRSPTESFVI RLENCHATAV GKIVTLTFK GTEEAALPGH LKVTGVNAGR LGIALLDTDG SSLLKPGTSH NKGQGEKVTG NSLELPFGAY VVATPEALRT K SVVPGDYE ATATFELTYR UniProtKB: Fimbrial adapter PapK |
-Macromolecule #4: P fimbria tip G-adhesin PapG-II
| Macromolecule | Name: P fimbria tip G-adhesin PapG-II / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.62057 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKWFPALLF SLCVSGESSA WNNIVFYSLG DVNSYQGGNV VITQRPQFIT SWRPGIATVT WNQCNGPGFA DGFWAYYREY IAWVVFPKK VMTQNGYPLF IEVHNKGSWS EENTGDNDSY FFLKGYKWDE RAFDAGNLCQ KPGETTRLTE KFDDIIFKVA L PADLPLGD ...String: MKKWFPALLF SLCVSGESSA WNNIVFYSLG DVNSYQGGNV VITQRPQFIT SWRPGIATVT WNQCNGPGFA DGFWAYYREY IAWVVFPKK VMTQNGYPLF IEVHNKGSWS EENTGDNDSY FFLKGYKWDE RAFDAGNLCQ KPGETTRLTE KFDDIIFKVA L PADLPLGD YSVKIPYTSG MQRHFASYLG ARFKIPYNVA KTLPRENEML FLFKNIGGCR PSAQSLEIKH GDLSINSANN HY AAQTLSV SCDVPANIRF MLLRNTTPTY SHGKKFSVGL GHGWDSIVSV NGVDTGETTM RWYKAGTQNL TIGSRLYGES SKI QPGVLS GSATLLMILP UniProtKB: P fimbria tip G-adhesin PapG-II |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 227396 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7lhg: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)