+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23294 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
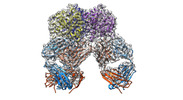
| Title | Trimeric human Arginase 1 in complex with mAb5 | |||||||||
 Map data Map data | Full map of 2x3hArg:3Mab5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Arginase / Metalloenzyme / IMMUNE SYSTEM / HYDROLASE-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of neutrophil mediated killing of fungus / Urea cycle / negative regulation of T-helper 2 cell cytokine production / arginase / arginase activity / : / urea cycle / response to nematode / defense response to protozoan / negative regulation of type II interferon-mediated signaling pathway ...positive regulation of neutrophil mediated killing of fungus / Urea cycle / negative regulation of T-helper 2 cell cytokine production / arginase / arginase activity / : / urea cycle / response to nematode / defense response to protozoan / negative regulation of type II interferon-mediated signaling pathway / negative regulation of activated T cell proliferation / L-arginine catabolic process / negative regulation of T cell proliferation / specific granule lumen / azurophil granule lumen / manganese ion binding / adaptive immune response / innate immune response / Neutrophil degranulation / extracellular space / extracellular region / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
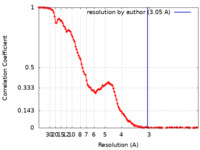
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Gomez-Llorente Y / Scapin G | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action. Authors: Rachel L Palte / Veronica Juan / Yacob Gomez-Llorente / Marc Andre Bailly / Kalyan Chakravarthy / Xun Chen / Daniel Cipriano / Ghassan N Fayad / Laurence Fayadat-Dilman / Symon Gathiaka / ...Authors: Rachel L Palte / Veronica Juan / Yacob Gomez-Llorente / Marc Andre Bailly / Kalyan Chakravarthy / Xun Chen / Daniel Cipriano / Ghassan N Fayad / Laurence Fayadat-Dilman / Symon Gathiaka / Heiko Greb / Brian Hall / Mas Handa / Mark Hsieh / Esther Kofman / Heping Lin / J Richard Miller / Nhung Nguyen / Jennifer O'Neil / Hussam Shaheen / Eric Sterner / Corey Strickland / Angie Sun / Shane Taremi / Giovanna Scapin /  Abstract: Human Arginase 1 (hArg1) is a metalloenzyme that catalyzes the hydrolysis of L-arginine to L-ornithine and urea, and modulates T-cell-mediated immune response. Arginase-targeted therapies have been ...Human Arginase 1 (hArg1) is a metalloenzyme that catalyzes the hydrolysis of L-arginine to L-ornithine and urea, and modulates T-cell-mediated immune response. Arginase-targeted therapies have been pursued across several disease areas including immunology, oncology, nervous system dysfunction, and cardiovascular dysfunction and diseases. Currently, all published hArg1 inhibitors are small molecules usually less than 350 Da in size. Here we report the cryo-electron microscopy structures of potent and inhibitory anti-hArg antibodies bound to hArg1 which form distinct macromolecular complexes that are greater than 650 kDa. With local resolutions of 3.5 Å or better we unambiguously mapped epitopes and paratopes for all five antibodies and determined that the antibodies act through orthosteric and allosteric mechanisms. These hArg1:antibody complexes present an alternative mechanism to inhibit hArg1 activity and highlight the ability to utilize antibodies as probes in the discovery and development of peptide and small molecule inhibitors for enzymes in general. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23294.map.gz emd_23294.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23294-v30.xml emd-23294-v30.xml emd-23294.xml emd-23294.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23294_fsc.xml emd_23294_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23294.png emd_23294.png | 76.6 KB | ||
| Masks |  emd_23294_msk_1.map emd_23294_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23294.cif.gz emd-23294.cif.gz | 6.6 KB | ||
| Others |  emd_23294_half_map_1.map.gz emd_23294_half_map_1.map.gz emd_23294_half_map_2.map.gz emd_23294_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23294 http://ftp.pdbj.org/pub/emdb/structures/EMD-23294 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23294 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23294 | HTTPS FTP |
-Validation report
| Summary document |  emd_23294_validation.pdf.gz emd_23294_validation.pdf.gz | 981 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23294_full_validation.pdf.gz emd_23294_full_validation.pdf.gz | 980.6 KB | Display | |
| Data in XML |  emd_23294_validation.xml.gz emd_23294_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_23294_validation.cif.gz emd_23294_validation.cif.gz | 23.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23294 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23294 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23294 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23294 | HTTPS FTP |
-Related structure data
| Related structure data |  7leyMC  7lexC  7lezC  7lf0C  7lf1C  7lf2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23294.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23294.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map of 2x3hArg:3Mab5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23294_msk_1.map emd_23294_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B.
| File | emd_23294_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A.
| File | emd_23294_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric human arginase in complex with mAb5
| Entire | Name: Trimeric human arginase in complex with mAb5 |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric human arginase in complex with mAb5
| Supramolecule | Name: Trimeric human arginase in complex with mAb5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: 2 trimers of human arginase, bound to 3 mAb5 molecules |
|---|---|
| Molecular weight | Theoretical: 427 KDa |
-Supramolecule #2: Arginase-1
| Supramolecule | Name: Arginase-1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: mAb5
| Supramolecule | Name: mAb5 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Arginase-1
| Macromolecule | Name: Arginase-1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: arginase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.779879 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAKSRTIGI IGAPFSKGQP RGGVEEGPTV LRKAGLLEKL KEQECDVKDY GDLPFADIPN DSPFQIVKNP RSVGKASEQL AGKVAEVKK NGRISLVLGG DHSLAIGSIS GHARVHPDLG VIWVDAHTDI NTPLTTTSGN LHGQPVSFLL KELKGKIPDV P GFSWVTPC ...String: MSAKSRTIGI IGAPFSKGQP RGGVEEGPTV LRKAGLLEKL KEQECDVKDY GDLPFADIPN DSPFQIVKNP RSVGKASEQL AGKVAEVKK NGRISLVLGG DHSLAIGSIS GHARVHPDLG VIWVDAHTDI NTPLTTTSGN LHGQPVSFLL KELKGKIPDV P GFSWVTPC ISAKDIVYIG LRDVDPGEHY ILKTLGIKYF SMTEVDRLGI GKVMEETLSY LLGRKKRPIH LSFDVDGLDP SF TPATGTP VVGGLTYREG LYITEEIYKT GLLSGLDIME VNPSLGKTPE EVTRTVNTAV AITLACFGLA REGNHKPIDY LNP PK UniProtKB: Arginase-1 |
-Macromolecule #2: mAb5 heavy chain
| Macromolecule | Name: mAb5 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.036074 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG VVRPGGSLRL SCAASGFTFD DYGMTWVRQA PGKGLEWVSG INWNGGSTGY ADSVKGRFTI SRDNAKNSLY LQMNSLRAE DTALYHCARD RRRGSYGSDA FDIWGQGTMV TVSSAKTTPP SVYPLAPGSA AQTNSMVTLG CLVKGYFPEP V TVTWNSGS ...String: EVQLVESGGG VVRPGGSLRL SCAASGFTFD DYGMTWVRQA PGKGLEWVSG INWNGGSTGY ADSVKGRFTI SRDNAKNSLY LQMNSLRAE DTALYHCARD RRRGSYGSDA FDIWGQGTMV TVSSAKTTPP SVYPLAPGSA AQTNSMVTLG CLVKGYFPEP V TVTWNSGS LSSGVHTFPA VLESDLYTLS SSVTVPSSPR PSETVTCNVA HPASSTKVDK KIVPRDCGCK PCICTVPEVS SV FIFPPKP KDVLTITLTP KVTCVVVDIS KDDPEVQFSW FVDDVEVHTA QTQPREEQFN STFRSVSELP IMHQDWLNGK EFK CRVNSA AFPAPIEKTI SKTKGRPKAP QVYTIPPPKE QMAKDKVSLT CMITDFFPED ITVEWQWNGQ PAENYKNTQP IMNT NGSYF VYSKLNVQKS NWEAGNTFTC SVLHEGLHNH HTEKSLSHSP GK |
-Macromolecule #3: mAb5 light chain
| Macromolecule | Name: mAb5 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.381787 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQGIS NYLAWYQQKP GKVPQLLISA ASTLQSGVPS RFSGSGSGTD FTLTISSLQP EDVATYYCQ KYNSAPRTFG QGTKVEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS ...String: DIQMTQSPSS LSASVGDRVT ITCRASQGIS NYLAWYQQKP GKVPQLLISA ASTLQSGVPS RFSGSGSGTD FTLTISSLQP EDVATYYCQ KYNSAPRTFG QGTKVEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS FNRNEC |
-Macromolecule #4: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Number real images: 1197 / Average exposure time: 6.0 sec. / Average electron dose: 44.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7ley: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)