[English] 日本語
 Yorodumi
Yorodumi- EMDB-23187: Mouse Norovirus Protruding domain complexed with neutralizing Fab... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23187 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse Norovirus Protruding domain complexed with neutralizing Fab fragment from mAb A6.2 | |||||||||
 Map data Map data | Soluble form of mouse norovirus protruding domain complexed with Fab fragments from neutralizing mAb A6.2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Antibody / norovirus / spike / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationantigenic variation / symbiont-mediated perturbation of host defense response / virion component / host cell cytoplasm Similarity search - Function | |||||||||
| Biological species |   Murine norovirus 1 / Murine norovirus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Smith TJ / Sherman MB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2021 Journal: J Virol / Year: 2021Title: A Norovirus Uses Bile Salts To Escape Antibody Recognition While Enhancing Receptor Binding. Authors: Alexis N Williams / Michael B Sherman / Hong Q Smith / Stefan Taube / B Montgomery Pettitt / Christiane E Wobus / Thomas J Smith /   Abstract: Noroviruses, members of the family, are the major cause of epidemic gastroenteritis in humans, causing ∼20 million cases annually. These plus-strand RNA viruses have T=3 icosahedral protein ...Noroviruses, members of the family, are the major cause of epidemic gastroenteritis in humans, causing ∼20 million cases annually. These plus-strand RNA viruses have T=3 icosahedral protein capsids with 90 pronounced protruding (P) domain dimers to which antibodies and cellular receptors bind. In the case of mouse norovirus (MNV), bile salts have been shown to enhance receptor (CD300lf) binding to the P domain. We demonstrated previously that the P domains of several genotypes are markedly flexible and "float" over the shell, but the role of this flexibility was unclear. Recently, we demonstrated that bile causes a 90° rotation and collapse of the P domain onto the shell surface. Since bile binds distally to the P-shell interface, it was not at all clear how it could cause such dramatic changes. Here, we present the near-atomic resolution cryo-electron microscopy (cryo-EM) structure of the MNV protruding domain complexed with a neutralizing Fab. On the basis of previous results, we show here that bile salts cause allosteric conformational changes in the P domain that block antibody recognition of the top of the P domain. In addition, bile causes a major rearrangement of the P domain dimers that is likely responsible for the bile-induced collapse of the P domain onto the shell. In the contracted shell conformation, antibodies to the P1 and shell domains are not expected to bind. Therefore, at the site of infection in the gut, the host's own bile allows the virus to escape antibody-mediated neutralization while enhancing cell attachment. The major feature of calicivirus capsids is the 90 protruding domains (P domains) that are the site of cell receptor attachment and antibody epitopes. We demonstrated previously that these P domains are highly mobile and that bile causes these "floating" P domains in mouse norovirus (MNV) to contract onto the shell surface. Here, we present the near-atomic cryo-EM structure of the isolated MNV P domain complexed with a neutralizing Fab fragment. Our data show that bile causes two sets of changes. First, bile causes allosteric conformational changes in the epitopes at the top of the P domain that block antibody binding. Second, bile causes the P domain dimer subunits to rotate relative to each other, causing a contraction of the P domain that buries epitopes at the base of the P and shell domains. Taken together, the results show that MNV uses the host's own metabolites to enhance cell receptor binding while simultaneously blocking antibody recognition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
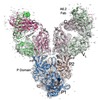
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23187.map.gz emd_23187.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23187-v30.xml emd-23187-v30.xml emd-23187.xml emd-23187.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23187.png emd_23187.png | 196.7 KB | ||
| Filedesc metadata |  emd-23187.cif.gz emd-23187.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23187 http://ftp.pdbj.org/pub/emdb/structures/EMD-23187 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23187 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23187 | HTTPS FTP |
-Validation report
| Summary document |  emd_23187_validation.pdf.gz emd_23187_validation.pdf.gz | 514.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23187_full_validation.pdf.gz emd_23187_full_validation.pdf.gz | 513.8 KB | Display | |
| Data in XML |  emd_23187_validation.xml.gz emd_23187_validation.xml.gz | 6.2 KB | Display | |
| Data in CIF |  emd_23187_validation.cif.gz emd_23187_validation.cif.gz | 7.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23187 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23187 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23187 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23187 | HTTPS FTP |
-Related structure data
| Related structure data |  7l5jMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23187.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23187.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Soluble form of mouse norovirus protruding domain complexed with Fab fragments from neutralizing mAb A6.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
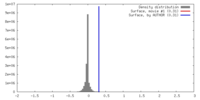
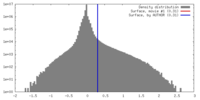
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Murine norovirus 1 - Fab complex
| Entire | Name: Murine norovirus 1 - Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: Murine norovirus 1 - Fab complex
| Supramolecule | Name: Murine norovirus 1 - Fab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Capsid protein
| Supramolecule | Name: Capsid protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Murine norovirus 1 Murine norovirus 1 |
-Supramolecule #3: Anti mouse norovirus mAb A6.2 Fab
| Supramolecule | Name: Anti mouse norovirus mAb A6.2 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Murine norovirus 1 Murine norovirus 1 |
| Molecular weight | Theoretical: 34.858281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNATIYRMVD LPVIQPRLCT HARWPAPVYG LLVDPSLPSN PQWQNGRVHV DGTLLGTTPI SGSWVSCFAA EAAYEFQSGT GEVATFTLI EQDGSAYVPG DRAAPLGYPD FSGQLEIEVQ TETTKTGDKL KVTTFEMILG PTTNADQAPY QGRVFASVTA A ASLDLVDG ...String: SNATIYRMVD LPVIQPRLCT HARWPAPVYG LLVDPSLPSN PQWQNGRVHV DGTLLGTTPI SGSWVSCFAA EAAYEFQSGT GEVATFTLI EQDGSAYVPG DRAAPLGYPD FSGQLEIEVQ TETTKTGDKL KVTTFEMILG PTTNADQAPY QGRVFASVTA A ASLDLVDG RVRAVPRSIY GFQDTIPEYN DGLLVPLAPP IGPFLPGEVL LRFRTYMRQI DTADAAAEAI DCALPQEFVS WF ASNAFTV QSEALLLRYR NTLTGQLLFE CKLYNEGYIA LSYSGSGPLT FPTDGIFEVV SWVPRLYQLA SVGSLATGRM LKQ UniProtKB: Capsid protein VP1 |
-Macromolecule #2: Anti mouse norovirus mAb A6.2 Fab light chain
| Macromolecule | Name: Anti mouse norovirus mAb A6.2 Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.083512 KDa |
| Sequence | String: QIVLTQSPAI MSASPGEKVT ITCSASSSVS YMHWFQQKPG TSPKLWIYST SNLASGVPAR FSGSGSGTSY SLTISRMEAE DAATYYCQQ RSSYPFTFGG GTKLEIKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK ...String: QIVLTQSPAI MSASPGEKVT ITCSASSSVS YMHWFQQKPG TSPKLWIYST SNLASGVPAR FSGSGSGTSY SLTISRMEAE DAATYYCQQ RSSYPFTFGG GTKLEIKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK DSTYSMSSTL TLTKDEYERH NSYTCEATHK TSTSPIVKSF NRN |
-Macromolecule #3: Anti mouse norovirus mAb A6.2 Fab heavy chain
| Macromolecule | Name: Anti mouse norovirus mAb A6.2 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.759061 KDa |
| Sequence | String: EVKLLESGGG LVQPGGSLKL SCAASGFDFS RYWMSWVRQA PGKGLEWIGQ INPHSSTINY TPSLRDKFII SRDNAKNTLY LQMTKVRSE DTALYYCARL LRYFYALDYW GQGASVTVSS AKTTPPSVYP LAPGRAAAAA SMVTLGCLVK GYFPEPVTVT W NSGSLAAG ...String: EVKLLESGGG LVQPGGSLKL SCAASGFDFS RYWMSWVRQA PGKGLEWIGQ INPHSSTINY TPSLRDKFII SRDNAKNTLY LQMTKVRSE DTALYYCARL LRYFYALDYW GQGASVTVSS AKTTPPSVYP LAPGRAAAAA SMVTLGCLVK GYFPEPVTVT W NSGSLAAG VHTFPAVLQA ALYTLSSSVT VPSSSWPSET VTCNVAHPAS STKVDKKIVP RAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)