+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23006 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of Hsp90 and co-chaperone p23 | |||||||||
 Map data Map data | Primary map ; Hsp90:p23 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationprostaglandin-E synthase / prostaglandin-E synthase activity / prostanoid biosynthetic process / Aryl hydrocarbon receptor signalling / telomerase activity / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / telomerase holoenzyme complex / prostaglandin biosynthetic process / telomerase holoenzyme complex assembly / : ...prostaglandin-E synthase / prostaglandin-E synthase activity / prostanoid biosynthetic process / Aryl hydrocarbon receptor signalling / telomerase activity / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / telomerase holoenzyme complex / prostaglandin biosynthetic process / telomerase holoenzyme complex assembly / : / protein folding chaperone complex / HSF1 activation / Attenuation phase / chaperone-mediated protein complex assembly / telomere maintenance via telomerase / positive regulation of telomere maintenance via telomerase / DNA polymerase binding / ESR-mediated signaling / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / telomere maintenance / Hsp90 protein binding / unfolded protein binding / protein-folding chaperone binding / protein folding / Estrogen-dependent gene expression / Potential therapeutics for SARS / chromosome, telomeric region / protein stabilization / signal transduction / protein-containing complex / nucleoplasm / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
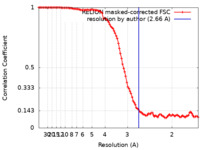
| Method | single particle reconstruction / cryo EM / Resolution: 2.66 Å | |||||||||
 Authors Authors | Noddings CM / Wang Y-R / Agard DA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structure of Hsp90-p23-GR reveals the Hsp90 client-remodelling mechanism. Authors: Chari M Noddings / Ray Yu-Ruei Wang / Jill L Johnson / David A Agard /  Abstract: Hsp90 is a conserved and essential molecular chaperone responsible for the folding and activation of hundreds of 'client' proteins. The glucocorticoid receptor (GR) is a model client that constantly ...Hsp90 is a conserved and essential molecular chaperone responsible for the folding and activation of hundreds of 'client' proteins. The glucocorticoid receptor (GR) is a model client that constantly depends on Hsp90 for activity. GR ligand binding was previously shown to nr inhibited by Hsp70 and restored by Hsp90, aided by the co-chaperone p23. However, a molecular understanding of the chaperone-mediated remodelling that occurs between the inactive Hsp70-Hsp90 'client-loading complex' and an activated Hsp90-p23 'client-maturation complex' is lacking for any client, including GR. Here we present a cryo-electron microscopy (cryo-EM) structure of the human GR-maturation complex (GR-Hsp90-p23), revealing that the GR ligand-binding domain is restored to a folded, ligand-bound conformation, while being simultaneously threaded through the Hsp90 lumen. In addition, p23 directly stabilizes native GR using a C-terminal helix, resulting in enhanced ligand binding. This structure of a client bound to Hsp90 in a native conformation contrasts sharply with the unfolded kinase-Hsp90 structure. Thus, aided by direct co-chaperone-client interactions, Hsp90 can directly dictate client-specific folding outcomes. Together with the GR-loading complex structure, we present the molecular mechanism of chaperone-mediated GR remodelling, establishing the first, to our knowledge, complete chaperone cycle for any Hsp90 client. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23006.map.gz emd_23006.map.gz | 6.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23006-v30.xml emd-23006-v30.xml emd-23006.xml emd-23006.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23006_fsc.xml emd_23006_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_23006.png emd_23006.png | 53.6 KB | ||
| Masks |  emd_23006_msk_1.map emd_23006_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_23006_additional_1.map.gz emd_23006_additional_1.map.gz emd_23006_half_map_1.map.gz emd_23006_half_map_1.map.gz emd_23006_half_map_2.map.gz emd_23006_half_map_2.map.gz | 80.3 MB 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23006 http://ftp.pdbj.org/pub/emdb/structures/EMD-23006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23006 | HTTPS FTP |
-Related structure data
| Related structure data |  7krjC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11028 (Title: Cryo-EM Structures of Glucocorticoid Receptor-Hsp90-p23 [the GR Maturation Complex], Hsp90-p23, and MBP-Hsp90-p23 EMPIAR-11028 (Title: Cryo-EM Structures of Glucocorticoid Receptor-Hsp90-p23 [the GR Maturation Complex], Hsp90-p23, and MBP-Hsp90-p23Data size: 494.1 Data #1: MotionCor2 aligned frames of GR-Hsp90-p23 collected on Gatan K3 [micrographs - single frame] Data #2: Processed subsets [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23006.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23006.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map ; Hsp90:p23 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23006_msk_1.map emd_23006_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Raw map ; Hsp90:p23
| File | emd_23006_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map ; Hsp90:p23 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 ; Hsp90:p23
| File | emd_23006_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 ; Hsp90:p23 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 ; Hsp90:p23
| File | emd_23006_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 ; Hsp90:p23 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Hsp90 dimer and the co-chaperone p23
| Entire | Name: Complex of Hsp90 dimer and the co-chaperone p23 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Hsp90 dimer and the co-chaperone p23
| Supramolecule | Name: Complex of Hsp90 dimer and the co-chaperone p23 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 188 KDa |
-Macromolecule #1: heat shock protein 90
| Macromolecule | Name: heat shock protein 90 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MPEETQTQDQ PMEEEEVETF AFQAEIAQLM SLIINTFYSN KEIFLRELIS NSSDALDKIR YESLTDPSKL DSGKELHINL IPNKQDRTLT IVDTGIGMTK ADLINNLGTI AKSGTKAFME ALQAGADISM IGQFGVGFYS AYLVAEKVTV ITKHNDDEQY AWESSAGGSF ...String: MPEETQTQDQ PMEEEEVETF AFQAEIAQLM SLIINTFYSN KEIFLRELIS NSSDALDKIR YESLTDPSKL DSGKELHINL IPNKQDRTLT IVDTGIGMTK ADLINNLGTI AKSGTKAFME ALQAGADISM IGQFGVGFYS AYLVAEKVTV ITKHNDDEQY AWESSAGGSF TVRTDTGEPM GRGTKVILHL KEDQTEYLEE RRIKEIVKKH SQFIGYPITL FVEKERDKEV SDDEAEEKED KEEEKEKEEK ESEDKPEIED VGSDEEEEKK DGDKKKKKKI KEKYIDQEEL NKTKPIWTRN PDDITNEEYG EFYKSLTNDW EDHLAVKHFS VEGQLEFRAL LFVPRRAPFD LFENRKKKNN IKLYVRRVFI MDNCEELIPE YLNFIRGVVD SEDLPLNISR EMLQQSKILK VIRKNLVKKC LELFTELAED KENYKKFYEQ FSKNIKLGIH EDSQNRKKLS ELLRYYTSAS GDEMVSLKDY CTRMKENQKH IYYITGETKD QVANSAFVER LRKHGLEVIY MIEPIDEYCV QQLKEFEGKT LVSVTKEGLE LPEDEEEKKK QEEKKTKFEN LCKIMKDILE KKVEKVVVSN RLVTSPCCIV TSTYGWTANM ERIMKAQALR DNSTMGYMAA KKHLEINPDH SIIETLRQKA EADKNDKSVK DLVILLYETA LLSSGFSLED PQTHANRIYR MIKLGLGIDE DDPTADDTSA AVTEEMPPLE GDDDTSRMEE VD |
-Macromolecule #2: p23
| Macromolecule | Name: p23 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MQPASAKWYD RRDYVFIEFC VEDSKDVNVN FEKSKLTFSC LGGSDNFKHL NEIDLFHCID PNDSKHKRTD RSILCCLRKG ESGQSWPRLT KERAKLNWLS VDFNNWKDWE DDSDEDMSNF DRFSEMMNNM GGDEDVDLPE VDGADDDSQD SDDEKMPDLE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 5608 / Average exposure time: 5.9 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)