[English] 日本語
 Yorodumi
Yorodumi- EMDB-22978: Structure of upper Tn Ca2+ free (rotated) and lower Tn Ca2+ bound... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22978 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of upper Tn Ca2+ free (rotated) and lower Tn Ca2+ bound cardiac native thin filament at pCa=5.8 | ||||||||||||
 Map data Map data | Structure of Ca2 free / Ca2 bound cardiac native thin filament at pCa=5.8 mode 1 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | actin / tropomyosin / troponin / TF / CONTRACTILE PROTEIN | ||||||||||||
| Biological species |  | ||||||||||||
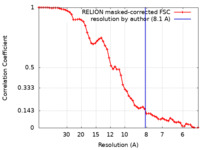
| Method | single particle reconstruction / cryo EM / Resolution: 8.1 Å | ||||||||||||
 Authors Authors | Galkin VE / Risi CM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: The structure of the native cardiac thin filament at systolic Ca levels. Authors: Cristina M Risi / Ian Pepper / Betty Belknap / Maicon Landim-Vieira / Howard D White / Kelly Dryden / Jose R Pinto / P Bryant Chase / Vitold E Galkin /  Abstract: Every heartbeat relies on cyclical interactions between myosin thick and actin thin filaments orchestrated by rising and falling Ca levels. Thin filaments are comprised of two actin strands, each ...Every heartbeat relies on cyclical interactions between myosin thick and actin thin filaments orchestrated by rising and falling Ca levels. Thin filaments are comprised of two actin strands, each harboring equally separated troponin complexes, which bind Ca to move tropomyosin cables away from the myosin binding sites and, thus, activate systolic contraction. Recently, structures of thin filaments obtained at low (pCa ∼9) or high (pCa ∼3) Ca levels revealed the transition between the Ca-free and Ca-bound states. However, in working cardiac muscle, Ca levels fluctuate at intermediate values between pCa ∼6 and pCa ∼7. The structure of the thin filament at physiological Ca levels is unknown. We used cryoelectron microscopy and statistical analysis to reveal the structure of the cardiac thin filament at systolic pCa = 5.8. We show that the two strands of the thin filament consist of a mixture of regulatory units, which are composed of Ca-free, Ca-bound, or mixed (e.g., Ca free on one side and Ca bound on the other side) troponin complexes. We traced troponin complex conformations along and across individual thin filaments to directly determine the structural composition of the cardiac native thin filament at systolic Ca levels. We demonstrate that the two thin filament strands are activated stochastically with short-range cooperativity evident only on one of the two strands. Our findings suggest a mechanism by which cardiac muscle is regulated by narrow range Ca fluctuations. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22978.map.gz emd_22978.map.gz | 15.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22978-v30.xml emd-22978-v30.xml emd-22978.xml emd-22978.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22978_fsc.xml emd_22978_fsc.xml | 5.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_22978.png emd_22978.png | 104.7 KB | ||
| Filedesc metadata |  emd-22978.cif.gz emd-22978.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22978 http://ftp.pdbj.org/pub/emdb/structures/EMD-22978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22978 | HTTPS FTP |
-Related structure data
| Related structure data |  7konMC  7ko4C  7ko5C  7ko7C  7korC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22978.map.gz / Format: CCP4 / Size: 16.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22978.map.gz / Format: CCP4 / Size: 16.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Ca2 free / Ca2 bound cardiac native thin filament at pCa=5.8 mode 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.712 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Structure of Ca2+ free / Ca2+ bound cardiac native thin filament ...
| Entire | Name: Structure of Ca2+ free / Ca2+ bound cardiac native thin filament at pCa=5.8 mode 2 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of Ca2+ free / Ca2+ bound cardiac native thin filament ...
| Supramolecule | Name: Structure of Ca2+ free / Ca2+ bound cardiac native thin filament at pCa=5.8 mode 2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.862613 KDa |
| Sequence | String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIITNWD DMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSGDGV T HNVPIYEG ...String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIITNWD DMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSGDGV T HNVPIYEG YALPHAIMRL DLAGRDLTDY LMKILTERGY SFVTTAEREI VRDIKEKLCY VALDFENEMA TAASSSSLEK SY ELPDGQV ITIGNERFRC PETLFQPSFI GMESAGIHET TYNSIMKCDI DIRKDLYANN VMSGGTTMYP GIADRMQKEI TAL APSTMK IKIIAPPERK YSVWIGGSIL ASLSTFQQMW ITKQEYDEAG PSIVHRKCF |
-Macromolecule #2: Tropomyosin alpha-1 chain
| Macromolecule | Name: Tropomyosin alpha-1 chain / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.921773 KDa |
| Sequence | String: ASMDAIKKKM QMLKLDKENA LDRAEQAEAD KKAAEDRSKQ LEDELVSLQK KLKGTEDELD KYSEALKDAQ EKLELAEKKA TDAEADVAS LNRRIQLVEE ELDRAQERLA TALQKLEEAE KAADESERGM KVIESRAQKD EEKMEIQEIQ LKEAKHIAED A DRKYEEVA ...String: ASMDAIKKKM QMLKLDKENA LDRAEQAEAD KKAAEDRSKQ LEDELVSLQK KLKGTEDELD KYSEALKDAQ EKLELAEKKA TDAEADVAS LNRRIQLVEE ELDRAQERLA TALQKLEEAE KAADESERGM KVIESRAQKD EEKMEIQEIQ LKEAKHIAED A DRKYEEVA RKLVIIESDL ERAEERAELS EGKCAELEEE LKTVTNNLKS LEAQAEKYSQ KEDRYEEEIK VLSDKLKEAE TR AEFAERS VTKLEKSIDD LEDELYAQKL KYKAISEELD HALNDMTSI |
-Macromolecule #3: Troponin T, cardiac muscle
| Macromolecule | Name: Troponin T, cardiac muscle / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.023039 KDa |
| Sequence | String: FDDIHRKRME KDLNELQALI EAHFENRKKE EEELVSLKDR IERRRAERAE QQRIRNEREK ERQNRLAEER ARREEEENRR KAEDEARKK KALSNMMHFG GYIQKQAQTE RKSGKRQTER EKKKKILAER RKVLAIDHLN EDQLREKAKE LWQSIYNLEA E KFDLQEKF KQQKYEINVL RNRINDNQ |
-Macromolecule #4: Troponin I, cardiac muscle
| Macromolecule | Name: Troponin I, cardiac muscle / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.639691 KDa |
| Sequence | String: ISASRKLQLK TLLLQIAKQE LEREAEERRG EKGRALSTRC QPLELAGLGF AELQDLCRQL HARVDKVDEE RYDIEAKVTK NITEIADLT QKIFDLRGKF KRPTLRRVRI SADAMMQALL GARAKESLDL RAHLKQVKKE DTEKENREVG DWRKNIDALS G MEGRKKKF ES |
-Macromolecule #5: Troponin C
| Macromolecule | Name: Troponin C / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.288287 KDa |
| Sequence | String: DDIYKAAVEQ LTEEQKNEFK AAFDIFVLGA EDGCISTKEL GKVMRMLGQN PTPEELQEMI DEVDEDGSGT VDFDEFLVMM VRCMKDDSK GKSEEELSDL FRMFDKNADG YIDLDELKIM LQATGETITE DDIEELMKDG DKNNDGRIDY DEFLEFMKGV E |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
| Details | non-helical filament |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 34.0 e/Å2 Details: Images were collected in movie-mode - 40 subframes at 0.85e/A^2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-7kon: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)