+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22429 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human HUWE1 (focused on WWE) | ||||||||||||||||||
 Map data Map data | map from refinement after classification focused on WWE | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of peroxisome proliferator activated receptor signaling pathway / histone ubiquitin ligase activity / negative regulation of mitochondrial fusion / protein branched polyubiquitination / positive regulation of type 2 mitophagy / HECT-type E3 ubiquitin transferase / : / ubiquitin-ubiquitin ligase activity / Golgi organization / protein monoubiquitination ...negative regulation of peroxisome proliferator activated receptor signaling pathway / histone ubiquitin ligase activity / negative regulation of mitochondrial fusion / protein branched polyubiquitination / positive regulation of type 2 mitophagy / HECT-type E3 ubiquitin transferase / : / ubiquitin-ubiquitin ligase activity / Golgi organization / protein monoubiquitination / protein K48-linked ubiquitination / positive regulation of protein ubiquitination / circadian regulation of gene expression / base-excision repair / protein polyubiquitination / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / Antigen processing: Ubiquitination & Proteasome degradation / ubiquitin-dependent protein catabolic process / secretory granule lumen / ficolin-1-rich granule lumen / proteasome-mediated ubiquitin-dependent protein catabolic process / membrane fusion / cell differentiation / positive regulation of canonical NF-kappaB signal transduction / Golgi membrane / Neutrophil degranulation / mitochondrion / DNA binding / RNA binding / extracellular exosome / extracellular region / nucleoplasm / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
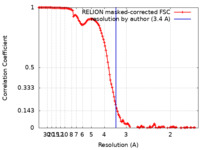
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||
 Authors Authors | Hunkeler M / Fischer ES | ||||||||||||||||||
| Funding support |  United States, United States,  Switzerland, 5 items Switzerland, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition. Authors: Moritz Hunkeler / Cyrus Y Jin / Michelle W Ma / Julie K Monda / Daan Overwijn / Eric J Bennett / Eric S Fischer /  Abstract: HECT ubiquitin ligases play essential roles in metazoan development and physiology. The HECT ligase HUWE1 is central to the cellular stress response by mediating degradation of key death or survival ...HECT ubiquitin ligases play essential roles in metazoan development and physiology. The HECT ligase HUWE1 is central to the cellular stress response by mediating degradation of key death or survival factors, including Mcl1, p53, DDIT4, and Myc. Although mutations in HUWE1 and related HECT ligases are widely implicated in human disease, our molecular understanding remains limited. Here we present a comprehensive investigation of full-length HUWE1, deepening our understanding of this class of enzymes. The N-terminal ∼3,900 amino acids of HUWE1 are indispensable for proper ligase function, and our cryo-EM structures of HUWE1 offer a complete molecular picture of this large HECT ubiquitin ligase. HUWE1 forms an alpha solenoid-shaped assembly with a central pore decorated with protein interaction modules. Structures of HUWE1 variants linked to neurodevelopmental disorders as well as of HUWE1 bound to a model substrate link the functions of this essential enzyme to its three-dimensional organization. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22429.map.gz emd_22429.map.gz | 14.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22429-v30.xml emd-22429-v30.xml emd-22429.xml emd-22429.xml | 32.1 KB 32.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22429_fsc.xml emd_22429_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_22429.png emd_22429.png | 35.9 KB | ||
| Masks |  emd_22429_msk_1.map emd_22429_msk_1.map | 184 MB |  Mask map Mask map | |
| Others |  emd_22429_additional_1.map.gz emd_22429_additional_1.map.gz emd_22429_additional_2.map.gz emd_22429_additional_2.map.gz emd_22429_additional_3.map.gz emd_22429_additional_3.map.gz emd_22429_half_map_1.map.gz emd_22429_half_map_1.map.gz emd_22429_half_map_2.map.gz emd_22429_half_map_2.map.gz | 162.5 MB 12.9 MB 11.4 MB 145.4 MB 145.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22429 http://ftp.pdbj.org/pub/emdb/structures/EMD-22429 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22429 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22429 | HTTPS FTP |
-Validation report
| Summary document |  emd_22429_validation.pdf.gz emd_22429_validation.pdf.gz | 510.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22429_full_validation.pdf.gz emd_22429_full_validation.pdf.gz | 510.3 KB | Display | |
| Data in XML |  emd_22429_validation.xml.gz emd_22429_validation.xml.gz | 21 KB | Display | |
| Data in CIF |  emd_22429_validation.cif.gz emd_22429_validation.cif.gz | 26.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22429 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22429 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22429 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22429 | HTTPS FTP |
-Related structure data
| Related structure data |  7mweMC  7jq9C  7mopC  7mwdC  7mwfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22429.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22429.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map from refinement after classification focused on WWE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
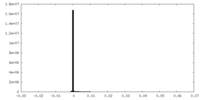
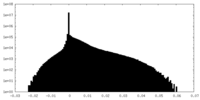
-Mask #1
| File |  emd_22429_msk_1.map emd_22429_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
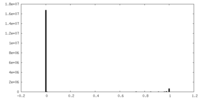
| Density Histograms |
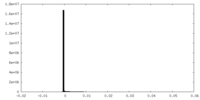
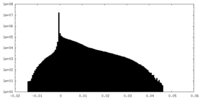
-Additional map: main map postprocessed with deepEMhancer
| File | emd_22429_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map postprocessed with deepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
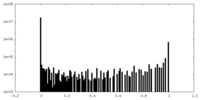
| Density Histograms |
-Additional map: main map blurred to B=-20A2
| File | emd_22429_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map blurred to B=-20A2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
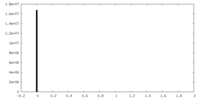
| Density Histograms |
-Additional map: main map blurred to B=10A2
| File | emd_22429_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map blurred to B=10A2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
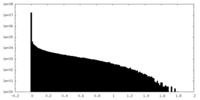
| Density Histograms |
-Half map: half map 1
| File | emd_22429_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_22429_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E3 ubiquitin-protein ligase HUWE1
| Entire | Name: E3 ubiquitin-protein ligase HUWE1 |
|---|---|
| Components |
|
-Supramolecule #1: E3 ubiquitin-protein ligase HUWE1
| Supramolecule | Name: E3 ubiquitin-protein ligase HUWE1 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: full length, crosslinked with BS3, refinement after classification focused on WWE domain |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 480 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant strain: Expi293 / Recombinant plasmid: pDEST Homo sapiens (human) / Recombinant strain: Expi293 / Recombinant plasmid: pDEST |
-Macromolecule #1: E3 ubiquitin-protein ligase HUWE1
| Macromolecule | Name: E3 ubiquitin-protein ligase HUWE1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: HECT-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKL AAANSSIDLI STSLYKKAGF KGTNSVDMKV DRTKLKKTPT EAPADCRALI DKLKVCNDEQ LLLELQQIKT WNIGKCELYH WVDLLDRFDG ILADAGQTVE NMSWMLVCDR PEREQLKMLL LAVLNFTALL IEYSFSRHLY SSIEHLTTLL ASSDMQVVLA ...String: MDYKDDDDKL AAANSSIDLI STSLYKKAGF KGTNSVDMKV DRTKLKKTPT EAPADCRALI DKLKVCNDEQ LLLELQQIKT WNIGKCELYH WVDLLDRFDG ILADAGQTVE NMSWMLVCDR PEREQLKMLL LAVLNFTALL IEYSFSRHLY SSIEHLTTLL ASSDMQVVLA VLNLLYVFSK RSNYITRLGS DKRTPLLTRL QHLAESWGGK ENGFGLAECC RDLHMMKYPP SATTLHFEFY ADPGAEVKIE KRTTSNTLHY IHIEQLDKIS ESPSEIMESL TKMYSIPKDK QMLLFTHIRL AHGFSNHRKR LQAVQARLHA ISILVYSNAL QESANSILYN GLIEELVDVL QITDKQLMEI KAASLRTLTS IVHLERTPKL SSIIDCTGTA SYHGFLPVLV RNCIQAMIDP SMDPYPHQFA TALFSFLYHL ASYDAGGEAL VSCGMMEALL KVIKFLGDEQ DQITFVTRAV RVVDLITNLD MAAFQSHSGL SIFIYRLEHE VDLCRKECPF VIKPKIQRPN TTQEGEEMET DMDGVQCIPQ RAALLKSMLN FLKKAIQDPA FSDGIRHVMD GSLPTSLKHI ISNAEYYGPS LFLLATEVVT VFVFQEPSLL SSLQDNGLTD VMLHALLIKD VPATREVLGS LPNVFSALCL NARGLQSFVQ CQPFERLFKV LLSPDYLPAM RRRRSSDPLG DTASNLGSAV DELMRHQPTL KTDATTAIIK LLEEICNLGR DPKYICQKPS IQKADGTATA PPPRSNHAAE EASSEDEEEE EVQAMQSFNS TQQNETEPNQ QVVGTEERIP IPLMDYILNV MKFVESILSN NTTDDHCQEF VNQKGLLPLV TILGLPNLPI DFPTSAACQA VAGVCKSILT LSHEPKVLQE GLLQLDSILS SLEPLHRPIE SPGGSVLLRE LACAGNVADA TLSAQATPLL HALTAAHAYI MMFVHTCRVG QSEIRSISVN QWGSQLGLSV LSKLSQLYCS LVWESTVLLS LCTPNSLPSG CEFGQADMQK LVPKDEKAGT TQGGKRSDGE QDGAAGSMDA STQGLLEGIG LDGDTLAPME TDEPTASDSK GKSKITPAMA ARIKQIKPLL SASSRLGRAL AELFGLLVKL CVGSPVRQRR SHHAASTTTA PTPAARSTAS ALTKLLTKGL SWQPPPYTPT PRFRLTFFIC SVGFTSPMLF DERKYPYHLM LQKFLCSGGH NALFETFNWA LSMGGKVPVS EGLEHSDLPD GTGEFLDAWL MLVEKMVNPT TVLESPHSLP AKLPGGVQNF PQFSALRFLV VTQKAAFTCI KNLWNRKPLK VYGGRMAESM LAILCHILRG EPVIRERLSK EKEGSRGEED TGQEEGGSRR EPQVNQQQLQ QLMDMGFTRE HAMEALLNTS TMEQATEYLL THPPPIMGGV VRDLSMSEED QMMRAIAMSL GQDIPMDQRA ESPEEVACRK EEEERKAREK QEEEEAKCLE KFQDADPLEQ DELHTFTDTM LPGCFHLLDE LPDTVYRVCD LIMTAIKRNG ADYRDMILKQ VVNQVWEAAD VLIKAALPLT TSDTKTVSEW ISQMATLPQA SNLATRILLL TLLFEELKLP CAWVVESSGI LNVLIKLLEV VQPCLQAAKE QKEVQTPKWI TPVLLLIDFY EKTAISSKRR AQMTKYLQSN SNNWRWFDDR SGRWCSYSAS NNSTIDSAWK SGETSVRFTA GRRRYTVQFT TMVQVNEETG NRRPVMLTLL RVPRLNKNSK NSNGQELEKT LEESKEMDIK RKENKGNDTP LALESTNTEK ETSLEETKIG EILIQGLTED MVTVLIRACV SMLGVPVDPD TLHATLRLCL RLTRDHKYAM MFAELKSTRM ILNLTQSSGF NGFTPLVTLL LRHIIEDPCT LRHTMEKVVR SAATSGAGST TSGVVSGSLG SREINYILRV LGPAACRNPD IFTEVANCCI RIALPAPRGS GTASDDEFEN LRIKGPNAVQ LVKTTPLKPS PLPVIPDTIK EVIYDMLNAL AAYHAPEEAD KSDPKPGVMT QEVGQLLQDM GDDVYQQYRS LTRQSSDFDT QSGFSINSQV FAADGASTET SASGTSQGEA STPEESRDGK KDKEGDRASE EGKQKGKGSK PLMPTSTILR LLAELVRSYV GIATLIANYS YTVGQSELIK EDCSVLAFVL DHLLPHTQNA EDKDTPALAR LFLASLAAAG SGTDAQVALV NEVKAALGRA LAMAESTEKH ARLQAVMCII STIMESCPST SSFYSSATAK TQHNGMNNII RLFLKKGLVN DLARVPHSLD LSSPNMANTV NAALKPLETL SRIVNQPSSL FGSKSASSKN KSEQDAQGAS QDSSSNQQDP GEPGEAEVQE EDHDVTQTEV ADGDIMDGEA ETDSVVIAGQ PEVLSSQEMQ VENELEDLID ELLERDGGSG NSTIIVSRSG EDESQEDVLM DEAPSNLSQA STLQANREDS MNILDPEDEE EHTQEEDSSG SNEDEDDSQD EEEEEEEDEE DDQEDDEGEE GDEDDDDDGS EMELDEDYPD MNASPLVRFE RFDREDDLII EFDNMFSSAT DIPPSPGNIP TTHPLMVRHA DHSSLTLGSG SSTTRLTQGI GRSQRTLRQL TANTGHTIHV HYPGNRQPNP PLILQRLLGP SAAADILQLS SSLPLQSRGR ARLLVGNDDV HIIARSDDEL LDDFFHDQST ATSQAGTLSS IPTALTRWTE ECKVLDAESM HDCVSVVKVS IVNHLEFLRD EELEERREKR RKQLAEEETK ITDKGKEDKE NRDQSAQCTA SKSNDSTEQN LSDGTPMPDS YPTTPSSTDA ATSESKETLG TLQSSQQQPT LPTPPALGEV PQELQSPAGE GGSSTQLLMP VEPEELGPTR PSGEAETTQM ELSPAPTITS LSPERAEDSD ALTAVSSQLE GSPMDTSSLA SCTLEEAVGD TSAAGSSEQP RAGSSTPGDA PPAVAEVQGR SDGSGESAQP PEDSSPPASS ESSSTRDSAV AISGADSRGI LEEPLPSTSS EEEDPLAGIS LPEGVDPSFL AALPDDIRRE VLQNQLGIRP PTRTAPSTNS SAPAVVGNPG VTEVSPEFLA ALPPAIQEEV LAQQRAEQQR RELAQNASSD TPMDPVTFIQ TLPSDLRRSV LEDMEDSVLA VMPPDIAAEA QALRREQEAR QRQLMHERLF GHSSTSALSA ILRSPAFTSR LSGNRGVQYT RLAVQRGGTF QMGGSSSHNR PSGSNVDTLL RLRGRLLLDH EALSCLLVLL FVDEPKLNTS RLHRVLRNLC YHAQTRHWVI RSLLSILQRS SESELCIETP KLTTSEEKGK KSSKSCGSSS HENRPLDLLH KMESKSSNQL SWLSVSMDAA LGCRTNIFQI QRSGGRKHTE KHASGGSTVH IHPQAAPVVC RHVLDTLIQL AKVFPSHFTQ QRTKETNCES DRERGNKACS PCSSQSSSSG ICTDFWDLLV KLDNMNVSRK GKNSVKSVPV SAGGEGETSP YSLEASPLGQ LMNMLSHPVI RRSSLLTEKL LRLLSLISIA LPENKVSEAQ ANSGSGASST TTATSTTSTT TTTAASTTPT PPTAPTPVTS APALVAATAI STIVVAASTT VTTPTTATTT VSISPTTKGS KSPAKVSDGG SSSTDFKMVS SGLTENQLQL SVEVLTSHSC SEEGLEDAAN VLLQLSRGDS GTRDTVLKLL LNGARHLGYT LCKQIGTLLA ELREYNLEQQ RRAQCETLSP DGLPEEQPQT TKLKGKMQSR FDMAENVVIV ASQKRPLGGR ELQLPSMSML TSKTSTQKFF LRVLQVIIQL RDDTRRANKK AKQTGRLGSS GLGSASSIQA AVRQLEAEAD AIIQMVREGQ RARRQQQAAT SESSQSEASV RREESPMDVD QPSPSAQDTQ SIASDGTPQG EKEKEERPPE LPLLSEQLSL DELWDMLGEC LKELEESHDQ HAVLVLQPAV EAFFLVHATE RESKPPVRDT RESQLAHIKD EPPPLSPAPL TPATPSSLDP FFSREPSSMH ISSSLPPDTQ KFLRFAETHR TVLNQILRQS TTHLADGPFA VLVDYIRVLD FDVKRKYFRQ ELERLDEGLR KEDMAVHVRR DHVFEDSYRE LHRKSPEEMK NRLYIVFEGE EGQDAGGLLR EWYMIISREM FNPMYALFRT SPGDRVTYTI NPSSHCNPNH LSYFKFVGRI VAKAVYDNRL LECYFTRSFY KHILGKSVRY TDMESEDYHF YQGLVYLLEN DVSTLGYDLT FSTEVQEFGV CEVRDLKPNG ANILVTEENK KEYVHLVCQM RMTGAIRKQL AAFLEGFYEI IPKRLISIFT EQELELLISG LPTIDIDDLK SNTEYHKYQS NSIQIQWFWR ALRSFDQADR AKFLQFVTGT SKVPLQGFAA LEGMNGIQKF QIHRDDRSTD RLPSAHTCFN QLDLPAYESF EKLRHMLLLA IQECSEGFGL A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: LEICA EM GP Details: CHAPSO detergent added to final conc. of 0.8 mM. Sample applied twice.. | |||||||||
| Details | Sample crosslinked with BS3. Monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Data colleciton in counting mode, using multi-shot scheme (4 holes per stage position, 3 movies per hole) |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 10390 / Average exposure time: 2.4 sec. / Average electron dose: 45.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)