[English] 日本語
 Yorodumi
Yorodumi- EMDB-22400: Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22400 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048 | |||||||||
 Map data Map data | Contour level 0.4 set in Chimera with step level 1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | intasome / integrase-viral DNA complex / HYDROLASE-DNA-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationribonuclease H / Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases ...ribonuclease H / Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / DNA-directed DNA polymerase activity / viral translational frameshifting / symbiont entry into host cell / proteolysis / DNA binding / zinc ion binding Similarity search - Function | |||||||||
| Biological species |  Rous sarcoma virus (strain Schmidt-Ruppin A) Rous sarcoma virus (strain Schmidt-Ruppin A) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Pandey KK / Bera S | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome. Authors: Krishan K Pandey / Sibes Bera / Ke Shi / Michael J Rau / Amarachi V Oleru / James A J Fitzpatrick / Alan N Engelman / Hideki Aihara / Duane P Grandgenett /  Abstract: Despite conserved catalytic integration mechanisms, retroviral intasomes composed of integrase (IN) and viral DNA possess diverse structures with variable numbers of IN subunits. To investigate ...Despite conserved catalytic integration mechanisms, retroviral intasomes composed of integrase (IN) and viral DNA possess diverse structures with variable numbers of IN subunits. To investigate intasome assembly mechanisms, we employed the Rous sarcoma virus (RSV) IN dimer that assembles a precursor tetrameric structure in transit to the mature octameric intasome. We determined the structure of RSV octameric intasome stabilized by a HIV-1 IN strand transfer inhibitor using single particle cryo-electron microscopy. The structure revealed significant flexibility of the two non-catalytic distal IN dimers along with previously unrecognized movement of the conserved intasome core, suggesting ordered conformational transitions between intermediates that may be important to capture the target DNA. Single amino acid substitutions within the IN C-terminal domain affected intasome assembly and function in vitro and infectivity of pseudotyped RSV virions. Unexpectedly, 17 C-terminal amino acids of IN were dispensable for virus infection despite regulating the transition of the tetrameric intasome to the octameric form in vitro. We speculate that this region may regulate the binding of highly flexible distal IN dimers to the intasome core to form the octameric complex. Our studies reveal key steps in the assembly of RSV intasomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22400.map.gz emd_22400.map.gz | 203.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22400-v30.xml emd-22400-v30.xml emd-22400.xml emd-22400.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22400_fsc.xml emd_22400_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_22400.png emd_22400.png | 72.8 KB | ||
| Filedesc metadata |  emd-22400.cif.gz emd-22400.cif.gz | 6.7 KB | ||
| Others |  emd_22400_half_map_1.map.gz emd_22400_half_map_1.map.gz emd_22400_half_map_2.map.gz emd_22400_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22400 http://ftp.pdbj.org/pub/emdb/structures/EMD-22400 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22400 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22400 | HTTPS FTP |
-Related structure data
| Related structure data |  7jn3MC  7ku7C  7kuiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22400.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22400.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Contour level 0.4 set in Chimera with step level 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half-map A: Contour level 0.4 set in Chimera with step level 1
| File | emd_22400_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map A: Contour level 0.4 set in Chimera with step level 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-map B: Contour level 0.4 set in Chimera with step level 1
| File | emd_22400_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map B: Contour level 0.4 set in Chimera with step level 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cleaved synaptic complex (CSC) formed with Rous sarcoma virus int...
| Entire | Name: Cleaved synaptic complex (CSC) formed with Rous sarcoma virus integrase and viral DNA in presence of HIV-1 integrase strand inhibitor MK-2048 |
|---|---|
| Components |
|
-Supramolecule #1: Cleaved synaptic complex (CSC) formed with Rous sarcoma virus int...
| Supramolecule | Name: Cleaved synaptic complex (CSC) formed with Rous sarcoma virus integrase and viral DNA in presence of HIV-1 integrase strand inhibitor MK-2048 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Rous sarcoma virus (strain Schmidt-Ruppin A) Rous sarcoma virus (strain Schmidt-Ruppin A) |
| Molecular weight | Theoretical: 257 KDa |
-Macromolecule #1: integrase
| Macromolecule | Name: integrase / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases |
|---|---|
| Source (natural) | Organism:  Rous sarcoma virus (strain Schmidt-Ruppin A) / Strain: Schmidt-Ruppin A Rous sarcoma virus (strain Schmidt-Ruppin A) / Strain: Schmidt-Ruppin A |
| Molecular weight | Theoretical: 30.926582 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PLREAKDLHT ALHIGPRALS KACNISMQQA REVVQTCPHC NSAPALEAGV NPRGLGPLQI WQTDFTLEPR MAPRSWLAVT VDTASSAIV VTQHGRVTSV AVQHHWATAI AVLGRPKAIK TDNGSCFTSK STREWLARWG IAHTTGIPGN SQGQAMVERA N RLLKDKIR ...String: PLREAKDLHT ALHIGPRALS KACNISMQQA REVVQTCPHC NSAPALEAGV NPRGLGPLQI WQTDFTLEPR MAPRSWLAVT VDTASSAIV VTQHGRVTSV AVQHHWATAI AVLGRPKAIK TDNGSCFTSK STREWLARWG IAHTTGIPGN SQGQAMVERA N RLLKDKIR VLAEGDGFMK RIPTSKQGEL LAKAMYALNH FERGENTKTP IQKHWRPTVL TEGPPVKIRI ETGEWEKGWN VL VWGRGYA AVKNRDTDKV IWVPSRKVKP DITQKDEVTK K UniProtKB: Gag-Pol polyprotein |
-Macromolecule #2: DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3')
| Macromolecule | Name: DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3') type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Rous sarcoma virus (strain Schmidt-Ruppin A) Rous sarcoma virus (strain Schmidt-Ruppin A) |
| Molecular weight | Theoretical: 5.5206 KDa |
| Sequence | String: (DA)(DA)(DT)(DG)(DT)(DT)(DG)(DT)(DC)(DT) (DT)(DA)(DT)(DG)(DC)(DA)(DA)(DT) |
-Macromolecule #3: DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3')
| Macromolecule | Name: DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3') type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Rous sarcoma virus (strain Schmidt-Ruppin A) Rous sarcoma virus (strain Schmidt-Ruppin A) |
| Molecular weight | Theoretical: 4.899232 KDa |
| Sequence | String: (DA)(DT)(DT)(DG)(DC)(DA)(DT)(DA)(DA)(DG) (DA)(DC)(DA)(DA)(DC)(DA) |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-...
| Macromolecule | Name: (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide type: ligand / ID: 5 / Number of copies: 2 / Formula: ZZX |
|---|---|
| Molecular weight | Theoretical: 461.874 Da |
| Chemical component information | 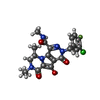 ChemComp-ZZX: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 5187 / Average electron dose: 66.0 e/Å2 Details: Images were collected in movie mode at 0.2 seconds per frame. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 30 / Target criteria: correlation coefficient |
| Output model |  PDB-7jn3: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)







































