+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22260 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of P5A-ATPase Spf1, Apo form | |||||||||
 Map data Map data | P5A-ATPase Spf1, Apo form | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | P-type ATPase / transmembrane helix dislocase / protein quality control / endoplasmic reticulum / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationextraction of mislocalized protein from ER membrane / membrane protein dislocase activity / sterol homeostasis / Ion transport by P-type ATPases / intracellular manganese ion homeostasis / Translocases; Catalysing the translocation of amino acids and peptides; Linked to the hydrolysis of a nucleoside triphosphate / P-type ion transporter activity / ATPase-coupled monoatomic cation transmembrane transporter activity / cis-Golgi network / protein hexamerization ...extraction of mislocalized protein from ER membrane / membrane protein dislocase activity / sterol homeostasis / Ion transport by P-type ATPases / intracellular manganese ion homeostasis / Translocases; Catalysing the translocation of amino acids and peptides; Linked to the hydrolysis of a nucleoside triphosphate / P-type ion transporter activity / ATPase-coupled monoatomic cation transmembrane transporter activity / cis-Golgi network / protein hexamerization / phosphatidylinositol-4-phosphate binding / protein unfolding / transmembrane transport / intracellular calcium ion homeostasis / protein transport / endoplasmic reticulum membrane / endoplasmic reticulum / ATP hydrolysis activity / mitochondrion / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
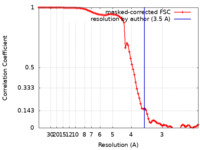
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Park E / Sim SI | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Authors: Michael J McKenna / Sue Im Sim / Alban Ordureau / Lianjie Wei / J Wade Harper / Sichen Shao / Eunyong Park /  Abstract: Organelle identity depends on protein composition. How mistargeted proteins are selectively recognized and removed from organelles is incompletely understood. Here, we found that the orphan P5A- ...Organelle identity depends on protein composition. How mistargeted proteins are selectively recognized and removed from organelles is incompletely understood. Here, we found that the orphan P5A-adenosine triphosphatase (ATPase) transporter ATP13A1 (Spf1 in yeast) directly interacted with the transmembrane segment (TM) of mitochondrial tail-anchored proteins. P5A-ATPase activity mediated the extraction of mistargeted proteins from the endoplasmic reticulum (ER). Cryo-electron microscopy structures of Spf1 revealed a large, membrane-accessible substrate-binding pocket that alternately faced the ER lumen and cytosol and an endogenous substrate resembling an α-helical TM. Our results indicate that the P5A-ATPase could dislocate misinserted hydrophobic helices flanked by short basic segments from the ER. TM dislocation by the P5A-ATPase establishes an additional class of P-type ATPase substrates and may correct mistakes in protein targeting or topogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22260.map.gz emd_22260.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22260-v30.xml emd-22260-v30.xml emd-22260.xml emd-22260.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22260_fsc.xml emd_22260_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22260.png emd_22260.png | 67.2 KB | ||
| Masks |  emd_22260_msk_1.map emd_22260_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22260.cif.gz emd-22260.cif.gz | 7 KB | ||
| Others |  emd_22260_additional.map.gz emd_22260_additional.map.gz emd_22260_half_map_1.map.gz emd_22260_half_map_1.map.gz emd_22260_half_map_2.map.gz emd_22260_half_map_2.map.gz | 59.7 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22260 http://ftp.pdbj.org/pub/emdb/structures/EMD-22260 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22260 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22260 | HTTPS FTP |
-Related structure data
| Related structure data |  6xmpMC  6xmqC  6xmsC  6xmtC  6xmuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22260.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22260.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | P5A-ATPase Spf1, Apo form | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.14 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22260_msk_1.map emd_22260_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: P5A-ATPase Spf1, Apo form
| File | emd_22260_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | P5A-ATPase Spf1, Apo form | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: P5A-ATPase Spf1, Apo form
| File | emd_22260_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | P5A-ATPase Spf1, Apo form | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: P5A-ATPase Spf1, Apo form
| File | emd_22260_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | P5A-ATPase Spf1, Apo form | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : P5A-ATPase Spf1 Apo form
| Entire | Name: P5A-ATPase Spf1 Apo form |
|---|---|
| Components |
|
-Supramolecule #1: P5A-ATPase Spf1 Apo form
| Supramolecule | Name: P5A-ATPase Spf1 Apo form / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: P5A-type ATPase
| Macromolecule | Name: P5A-type ATPase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of inorganic cations; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 137.573156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKKSFVSSP IVRDSTLLVP KSLIAKPYVL PFFPLYATFA QLYFQQYDRY IKGPEWTFVY LGTLVSLNIL VMLMPAWNVK IKAKFNYST TKNVNEATHI LIYTTPNNGS DGIVEIQRVT EAGSLQTFFQ FQKKRFLWHE NEQVFSSPKF LVDESPKIGD F QKCKGHSG ...String: MTKKSFVSSP IVRDSTLLVP KSLIAKPYVL PFFPLYATFA QLYFQQYDRY IKGPEWTFVY LGTLVSLNIL VMLMPAWNVK IKAKFNYST TKNVNEATHI LIYTTPNNGS DGIVEIQRVT EAGSLQTFFQ FQKKRFLWHE NEQVFSSPKF LVDESPKIGD F QKCKGHSG DLTHLKRLYG ENSFDIPIPT FMELFKEHAV APLFVFQVFC VALWLLDEFW YYSLFNLFMI ISMEAAAVFQ RL TALKEFR TMGIKPYTIN VFRNKKWVAL QTNELLPMDL VSITRTAEES AIPCDLILLD GSAIVNEAML SGESTPLLKE SIK LRPSED NLQLDGVDKI AVLHGGTKAL QVTPPEHKSD IPPPPDGGAL AIVTKTGFET SQGSLVRVMI YSAERVSVDN KEAL MFILF LLIFAVIASW YVWVEGTKMG RIQSKLILDC ILIITSVVPP ELPMELTMAV NSSLAALAKF YVYCTEPFRI PFAGR IDVC CFDKTGTLTG EDLVFEGLAG ISADSENIRH LYSAAEAPES TILVIGAAHA LVKLEDGDIV GDPMEKATLK AVGWAV ERK NSNYREGTGK LDIIRRFQFS SALKRSASIA SHNDALFAAV KGAPETIRER LSDIPKNYDE IYKSFTRSGS RVLALAS KS LPKMSQSKID DLNRDDVESE LTFNGFLIFH CPLKDDAIET IKMLNESSHR SIMITGDNPL TAVHVAKEVG IVFGETLI L DRAGKSDDNQ LLFRDVEETV SIPFDPSKDT FDHSKLFDRY DIAVTGYALN ALEGHSQLRD LLRHTWVYAR VSPSQKEFL LNTLKDMGYQ TLMCGDGTND VGALKQAHVG IALLNGTEEG LKKLGEQRRL EGMKMMYIKQ TEFMARWNQP QPPVPEPIAH LFPPGPKNP HYLKALESKG TVITPEIRKA VEEANSKPVE VIKPNGLSEK KPADLASLLL NSAGDAQGDE APALKLGDAS C AAPFTSKL ANVSAVTNII RQGRCALVNT IQMYKILALN CLISAYSLSI IYMAGVKFGD GQATVSGLLL SVCFLSISRG KP LEKLSKQ RPQSGIFNVY IMGSILSQFA VHIATLVYIT TEIYKLEPRE PQVDLEKEFA PSLLNTGIFI IQLVQQVSTF AVN YQGEPF RENIRSNKGM YYGLLGVTGL ALASATEFLP ELNEAMKFVP MTDDFKIKLT LTLLLDFFGS WGVEHFFKFF FMDD KPSDI SVQQVKIASK GATGGSTAGG ATTASGTGEN LYFQ UniProtKB: Endoplasmic reticulum transmembrane helix translocase |
-Macromolecule #2: DODECYL-BETA-D-MALTOSIDE
| Macromolecule | Name: DODECYL-BETA-D-MALTOSIDE / type: ligand / ID: 2 / Number of copies: 12 / Formula: LMT |
|---|---|
| Molecular weight | Theoretical: 510.615 Da |
| Chemical component information |  ChemComp-LMT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 35 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)