+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22168 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of EBV BFLF1 | |||||||||
 Map data Map data | BFLF1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | viral packaging / viral cleavage / VIRAL PROTEIN | |||||||||
| Function / homology | Herpesvirus major envelope glycoprotein / Herpesvirus putative major envelope glycoprotein / Herpesviridae UL32 packaging protein family profile. / host cell cytoplasm / viral envelope / host cell nucleus / metal ion binding / Packaging protein UL32 / Packaging protein UL32 homolog Function and homology information Function and homology information | |||||||||
| Biological species |  Epstein-Barr virus (strain GD1) (Epstein-Barr virus) Epstein-Barr virus (strain GD1) (Epstein-Barr virus) | |||||||||
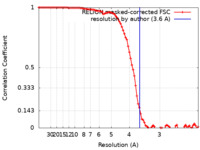
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Didychuk AL / Gates SN / Martin A / Glaunsinger B | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: A pentameric protein ring with novel architecture is required for herpesviral packaging. Authors: Allison L Didychuk / Stephanie N Gates / Matthew R Gardner / Lisa M Strong / Andreas Martin / Britt A Glaunsinger /  Abstract: Genome packaging in large double-stranded DNA viruses requires a powerful molecular motor to force the viral genome into nascent capsids, which involves essential accessory factors that are poorly ...Genome packaging in large double-stranded DNA viruses requires a powerful molecular motor to force the viral genome into nascent capsids, which involves essential accessory factors that are poorly understood. Here, we present structures of two such accessory factors from the oncogenic herpesviruses Kaposi's sarcoma-associated herpesvirus (KSHV; ORF68) and Epstein-Barr virus (EBV; BFLF1). These homologous proteins form highly similar homopentameric rings with a positively charged central channel that binds double-stranded DNA. Mutation of individual positively charged residues within but not outside the channel ablates DNA binding, and in the context of KSHV infection, these mutants fail to package the viral genome or produce progeny virions. Thus, we propose a model in which ORF68 facilitates the transfer of newly replicated viral genomes to the packaging motor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22168.map.gz emd_22168.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22168-v30.xml emd-22168-v30.xml emd-22168.xml emd-22168.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22168_fsc.xml emd_22168_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_22168.png emd_22168.png | 328.8 KB | ||
| Filedesc metadata |  emd-22168.cif.gz emd-22168.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22168 http://ftp.pdbj.org/pub/emdb/structures/EMD-22168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22168 | HTTPS FTP |
-Related structure data
| Related structure data |  6xfaMC  6xf9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22168.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22168.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BFLF1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.14 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Decamer structure of EBV BFLF1
| Entire | Name: Decamer structure of EBV BFLF1 |
|---|---|
| Components |
|
-Supramolecule #1: Decamer structure of EBV BFLF1
| Supramolecule | Name: Decamer structure of EBV BFLF1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Epstein-Barr virus (strain GD1) (Epstein-Barr virus) Epstein-Barr virus (strain GD1) (Epstein-Barr virus) |
| Molecular weight | Theoretical: 536 KDa |
-Macromolecule #1: Packaging protein UL32
| Macromolecule | Name: Packaging protein UL32 / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Epstein-Barr virus (strain GD1) (Epstein-Barr virus) Epstein-Barr virus (strain GD1) (Epstein-Barr virus)Strain: GD1 |
| Molecular weight | Theoretical: 53.678797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEDFVPWTVD NLKSQFEAVG LLMAHSYLPA NAEEGIAYPP LVHTYESLSP ASTCRVCDLL DTLVNHSDAP VAFFEDYALL CYYCLNAPR AWISSLITGM DFLHILIKYF PMAGGLDSLF MPSRILAIDI QLHFYICRCF LPVSSSDMIR NANLGYYKLE F LKSILTGQ ...String: MEDFVPWTVD NLKSQFEAVG LLMAHSYLPA NAEEGIAYPP LVHTYESLSP ASTCRVCDLL DTLVNHSDAP VAFFEDYALL CYYCLNAPR AWISSLITGM DFLHILIKYF PMAGGLDSLF MPSRILAIDI QLHFYICRCF LPVSSSDMIR NANLGYYKLE F LKSILTGQ SPANFCFKSM WPRTTPTFLT LPGPRTCKDS QDVPGDVGRG LYTALCCHLP TRNRVQHPFL RAEKGGLSPE IT TKADYCG LLLGTWQGTD LLGGPGHHAI GLNAEYSGDE LAELALAITR PEAGDHSQGP CLLAPMFGLR HKNASRTICP LCE SLGAHP DAKDTLDRFK SLILDSFGNN IKILDRIVFL IKTQNTLLDV PCPRLRAWLQ MCTPQDFHKH LFCDPLCAIN HSIT NPSVL FGQIYPPSFQ AFKAALAAGQ NLEQGVCDSL ITLVYIFKST QVARVGKTIL VDVTKELDVV LRIHGLDLVQ SYQTS QVYV UniProtKB: Packaging protein UL32 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 20 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||||||||
| Grid | Model: C-flat / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20 | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: 2 second blot, 3 second wait. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 839 / Average exposure time: 2.4 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)