+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22163 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AMC018 SOSIPv4.2 in complex with mAb RI808 | |||||||||
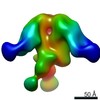
 Map data Map data | AMC018 SOSIPv4.2 in complex with mAb RI808 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 / unidentified (others) Human immunodeficiency virus 1 / unidentified (others) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Cottrell CA / Copps J / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Mining HIV controllers for broad and functional antibodies to recognize and eliminate HIV-infected cells. Authors: Evan D Rossignol / Anne-Sophie Dugast / Hacheming Compere / Christopher A Cottrell / Jeffrey Copps / Shu Lin / Deniz Cizmeci / Michael S Seaman / Margaret E Ackerman / Andrew B Ward / Galit Alter / Boris Julg /  Abstract: HIV monoclonal antibodies for viral reservoir eradication strategies will likely need to recognize reactivated infected cells and potently drive Fc-mediated innate effector cell activity. We ...HIV monoclonal antibodies for viral reservoir eradication strategies will likely need to recognize reactivated infected cells and potently drive Fc-mediated innate effector cell activity. We systematically characterize a library of 185 HIV-envelope-specific antibodies derived from 15 spontaneous HIV controllers (HCs) that selectively exhibit robust serum Fc functionality and compared them to broadly neutralizing antibodies (bNAbs) in clinical development. Within the 10 antibodies with the broadest cell-recognition capability, seven originated from HCs and three were bNAbs. V3-loop-targeting antibodies are enriched among the top cell binders, suggesting the V3-loop may be selectively exposed and accessible on the cell surface. Fc functionality is more variable across antibodies, which is likely influenced by distinct binding topology and corresponding Fc accessibility, highlighting not only the importance of target-cell recognition but also the need to optimize for Fc-mediated elimination. Ultimately, our results demonstrate that this comprehensive selection process can identify monoclonal antibodies poised to eliminate infected cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22163.map.gz emd_22163.map.gz | 11.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22163-v30.xml emd-22163-v30.xml emd-22163.xml emd-22163.xml | 9.1 KB 9.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22163.png emd_22163.png | 49.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22163 http://ftp.pdbj.org/pub/emdb/structures/EMD-22163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22163 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22163.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22163.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AMC018 SOSIPv4.2 in complex with mAb RI808 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AMC018 SOSIPv4.2 in complex with mAb RI808
| Entire | Name: AMC018 SOSIPv4.2 in complex with mAb RI808 |
|---|---|
| Components |
|
-Supramolecule #1: AMC018 SOSIPv4.2 in complex with mAb RI808
| Supramolecule | Name: AMC018 SOSIPv4.2 in complex with mAb RI808 / type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: AMC018 SOSIPv4.2
| Supramolecule | Name: AMC018 SOSIPv4.2 / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Supramolecule #3: mAb RI808
| Supramolecule | Name: mAb RI808 / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
| Grid | Details: unspecified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 26.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 7954 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















