[English] 日本語
 Yorodumi
Yorodumi- EMDB-8693: BG505 SOSIP.664 in complex with broadly neutralizing antibodies B... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8693 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
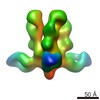
| Title | BG505 SOSIP.664 in complex with broadly neutralizing antibodies BG1 and 8ANC195 | ||||||||||||
 Map data Map data | cryo-EM map of BG505 SOSIP.664 in complex with broadly neutralizing antibodies BG1 and 8ANC195 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV-1 Env / broadly neutralizing antibodies / cryo-EM / single particle analysis / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationIgD immunoglobulin complex / IgA immunoglobulin complex / IgM immunoglobulin complex / IgE immunoglobulin complex / CD22 mediated BCR regulation / Fc epsilon receptor (FCERI) signaling / IgG immunoglobulin complex / Classical antibody-mediated complement activation / Initial triggering of complement / immunoglobulin mediated immune response ...IgD immunoglobulin complex / IgA immunoglobulin complex / IgM immunoglobulin complex / IgE immunoglobulin complex / CD22 mediated BCR regulation / Fc epsilon receptor (FCERI) signaling / IgG immunoglobulin complex / Classical antibody-mediated complement activation / Initial triggering of complement / immunoglobulin mediated immune response / FCGR activation / Role of LAT2/NTAL/LAB on calcium mobilization / symbiont-mediated perturbation of host defense response / Role of phospholipids in phagocytosis / immunoglobulin complex / Scavenging of heme from plasma / antigen binding / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / FCERI mediated Ca+2 mobilization / FCGR3A-mediated IL10 synthesis / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / host cell endosome membrane / Regulation of Complement cascade / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / FCGR3A-mediated phagocytosis / FCERI mediated MAPK activation / Regulation of actin dynamics for phagocytic cup formation / FCERI mediated NF-kB activation / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / blood microparticle / clathrin-dependent endocytosis of virus by host cell / Potential therapeutics for SARS / adaptive immune response / immune response / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / extracellular space / extracellular exosome / extracellular region / metal ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | ||||||||||||
 Authors Authors | Wang H / Bjorkman PJ | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Asymmetric recognition of HIV-1 Envelope trimer by V1V2 loop-targeting antibodies. Authors: Haoqing Wang / Harry B Gristick / Louise Scharf / Anthony P West / Rachel P Galimidi / Michael S Seaman / Natalia T Freund / Michel C Nussenzweig / Pamela J Bjorkman /  Abstract: The HIV-1 envelope (Env) glycoprotein binds to host cell receptors to mediate membrane fusion. The prefusion Env trimer is stabilized by V1V2 loops that interact at the trimer apex. Broadly ...The HIV-1 envelope (Env) glycoprotein binds to host cell receptors to mediate membrane fusion. The prefusion Env trimer is stabilized by V1V2 loops that interact at the trimer apex. Broadly neutralizing antibodies (bNAbs) against V1V2 loops, exemplified by PG9, bind asymmetrically as a single Fab to the apex of the symmetric Env trimer using a protruding CDRH3 to penetrate the Env glycan shield. Here we characterized a distinct mode of V1V2 epitope recognition by the new bNAb BG1 in which two Fabs bind asymmetrically per Env trimer using a compact CDRH3. Comparisons between cryo-EM structures of Env trimer complexed with BG1 (6.2 Å resolution) and PG9 (11.5 Å resolution) revealed a new V1V2-targeting strategy by BG1. Analyses of the EM structures provided information relevant to vaccine design including molecular details for different modes of asymmetric recognition of Env trimer and a binding model for BG1 recognition of V1V2 involving glycan flexibility. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8693.map.gz emd_8693.map.gz | 46.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8693-v30.xml emd-8693-v30.xml emd-8693.xml emd-8693.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8693_fsc.xml emd_8693_fsc.xml | 8.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_8693.png emd_8693.png | 73.4 KB | ||
| Filedesc metadata |  emd-8693.cif.gz emd-8693.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8693 http://ftp.pdbj.org/pub/emdb/structures/EMD-8693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8693 | HTTPS FTP |
-Related structure data
| Related structure data |  5viyMC  8695C  5vj6C  5vvfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8693.map.gz / Format: CCP4 / Size: 51.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8693.map.gz / Format: CCP4 / Size: 51.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map of BG505 SOSIP.664 in complex with broadly neutralizing antibodies BG1 and 8ANC195 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : BG505 SOSIP-BG1-8ANC195 complex
+Supramolecule #1: BG505 SOSIP-BG1-8ANC195 complex
+Supramolecule #2: BG505 SOSIP
+Supramolecule #3: BG1
+Supramolecule #4: 8ANC195
+Macromolecule #1: Envelope glycoprotein gp160
+Macromolecule #2: Envelope glycoprotein gp160
+Macromolecule #3: BG1 Fab light chain
+Macromolecule #4: BG1 Fab heavy chain
+Macromolecule #5: 8ANC195 G52K5 Fab heavy chain
+Macromolecule #6: 8ANC195 G52K5 Fab light chain
+Macromolecule #15: 2-acetamido-2-deoxy-beta-D-glucopyranose
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5viy: |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























