[English] 日本語
 Yorodumi
Yorodumi- EMDB-22106: High resolution cryoEM structure of huntingtin in complex with HAP40 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22106 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | High resolution cryoEM structure of huntingtin in complex with HAP40 | |||||||||
 Map data Map data | relion local resolution filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Huntingtin / HTT / 40-kDa huntingtin-associated protein / HAP40 / Structural Genomics / Structural Genomics Consortium / SGC / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationvesicle cytoskeletal trafficking / : / positive regulation of CAMKK-AMPK signaling cascade / microtubule-based transport / vocal learning / negative regulation of proteasomal protein catabolic process / regulation of CAMKK-AMPK signaling cascade / positive regulation of mitophagy / profilin binding / positive regulation of cilium assembly ...vesicle cytoskeletal trafficking / : / positive regulation of CAMKK-AMPK signaling cascade / microtubule-based transport / vocal learning / negative regulation of proteasomal protein catabolic process / regulation of CAMKK-AMPK signaling cascade / positive regulation of mitophagy / profilin binding / positive regulation of cilium assembly / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / vesicle transport along microtubule / positive regulation of aggrephagy / positive regulation of lipophagy / Golgi organization / dynein intermediate chain binding / dynactin binding / establishment of mitotic spindle orientation / phosphoprotein phosphatase activity / Regulation of MECP2 expression and activity / postsynaptic cytosol / beta-tubulin binding / presynaptic cytosol / heat shock protein binding / inclusion body / centriole / autophagosome / cytoplasmic vesicle membrane / negative regulation of extrinsic apoptotic signaling pathway / protein destabilization / kinase binding / p53 binding / late endosome / transmembrane transporter binding / early endosome / nuclear body / positive regulation of apoptotic process / axon / apoptotic process / dendrite / perinuclear region of cytoplasm / endoplasmic reticulum / Golgi apparatus / protein-containing complex / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
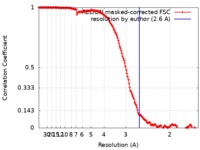
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Harding RJ / Deme JC / Lea SM / Arrowsmith CH / Structural Genomics Consortium (SGC) | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1. Authors: Rachel J Harding / Justin C Deme / Johannes F Hevler / Sem Tamara / Alexander Lemak / Jeffrey P Cantle / Magdalena M Szewczyk / Nola Begeja / Siobhan Goss / Xiaobing Zuo / Peter Loppnau / ...Authors: Rachel J Harding / Justin C Deme / Johannes F Hevler / Sem Tamara / Alexander Lemak / Jeffrey P Cantle / Magdalena M Szewczyk / Nola Begeja / Siobhan Goss / Xiaobing Zuo / Peter Loppnau / Alma Seitova / Ashley Hutchinson / Lixin Fan / Ray Truant / Matthieu Schapira / Jeffrey B Carroll / Albert J R Heck / Susan M Lea / Cheryl H Arrowsmith /     Abstract: Huntington's disease results from expansion of a glutamine-coding CAG tract in the huntingtin (HTT) gene, producing an aberrantly functioning form of HTT. Both wildtype and disease-state HTT form a ...Huntington's disease results from expansion of a glutamine-coding CAG tract in the huntingtin (HTT) gene, producing an aberrantly functioning form of HTT. Both wildtype and disease-state HTT form a hetero-dimer with HAP40 of unknown functional relevance. We demonstrate in vivo and in cell models that HTT and HAP40 cellular abundance are coupled. Integrating data from a 2.6 Å cryo-electron microscopy structure, cross-linking mass spectrometry, small-angle X-ray scattering, and modeling, we provide a near-atomic-level view of HTT, its molecular interaction surfaces and compacted domain architecture, orchestrated by HAP40. Native mass spectrometry reveals a remarkably stable hetero-dimer, potentially explaining the cellular inter-dependence of HTT and HAP40. The exon 1 region of HTT is dynamic but shows greater conformational variety in the polyglutamine expanded mutant than wildtype exon 1. Our data provide a foundation for future functional and drug discovery studies targeting Huntington's disease and illuminate the structural consequences of HTT polyglutamine expansion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22106.map.gz emd_22106.map.gz | 200.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22106-v30.xml emd-22106-v30.xml emd-22106.xml emd-22106.xml | 30.5 KB 30.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22106_fsc.xml emd_22106_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_22106.png emd_22106.png | 39.3 KB | ||
| Masks |  emd_22106_msk_1.map emd_22106_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22106.cif.gz emd-22106.cif.gz | 9.2 KB | ||
| Others |  emd_22106_additional_1.map.gz emd_22106_additional_1.map.gz emd_22106_additional_2.map.gz emd_22106_additional_2.map.gz emd_22106_half_map_1.map.gz emd_22106_half_map_1.map.gz emd_22106_half_map_2.map.gz emd_22106_half_map_2.map.gz | 320.2 MB 275 MB 276 MB 275.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22106 http://ftp.pdbj.org/pub/emdb/structures/EMD-22106 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22106 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22106 | HTTPS FTP |
-Validation report
| Summary document |  emd_22106_validation.pdf.gz emd_22106_validation.pdf.gz | 861.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22106_full_validation.pdf.gz emd_22106_full_validation.pdf.gz | 861.2 KB | Display | |
| Data in XML |  emd_22106_validation.xml.gz emd_22106_validation.xml.gz | 23.6 KB | Display | |
| Data in CIF |  emd_22106_validation.cif.gz emd_22106_validation.cif.gz | 31.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22106 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22106 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22106 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22106 | HTTPS FTP |
-Related structure data
| Related structure data |  6x9oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22106.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22106.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion local resolution filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
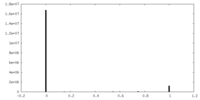
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22106_msk_1.map emd_22106_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
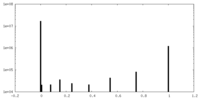
| Projections & Slices |
| ||||||||||||
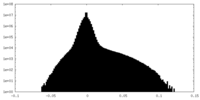
| Density Histograms |
-Additional map: relion postprocessed map
| File | emd_22106_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion postprocessed map | ||||||||||||
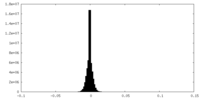
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: relion auto-refinement map
| File | emd_22106_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion auto-refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: relion auto-refinement unfiltered half map 2
| File | emd_22106_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion auto-refinement unfiltered half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: relion auto-refinement unfiltered half map 1
| File | emd_22106_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion auto-refinement unfiltered half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human huntingtin-HAP40 complex
| Entire | Name: Human huntingtin-HAP40 complex |
|---|---|
| Components |
|
-Supramolecule #1: Human huntingtin-HAP40 complex
| Supramolecule | Name: Human huntingtin-HAP40 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Huntingtin
| Macromolecule | Name: Huntingtin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 349.472344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATLEKLMKA FESLKSFQQQ QQQQQQQQQQ QQQQQQQQQQ PPPPPPPPPP PQLPQPPPQA QPLLPQPQPP PPPPPPPPGP AVAEEPLHR PKKELSATKK DRVNHCLTIC ENIVAQSVRN SPEFQKLLGI AMELFLLCSD DAESDVRMVA DECLNKVIKA L MDSNLPRL ...String: MATLEKLMKA FESLKSFQQQ QQQQQQQQQQ QQQQQQQQQQ PPPPPPPPPP PQLPQPPPQA QPLLPQPQPP PPPPPPPPGP AVAEEPLHR PKKELSATKK DRVNHCLTIC ENIVAQSVRN SPEFQKLLGI AMELFLLCSD DAESDVRMVA DECLNKVIKA L MDSNLPRL QLELYKEIKK NGAPRSLRAA LWRFAELAHL VRPQKCRPYL VNLLPCLTRT SKRPEESVQE TLAAAVPKIM AS FGNFAND NEIKVLLKAF IANLKSSSPT IRRTAAGSAV SICQHSRRTQ YFYSWLLNVL LGLLVPVEDE HSTLLILGVL LTL RYLVPL LQQQVKDTSL KGSFGVTRKE MEVSPSAEQL VQVYELTLHH TQHQDHNVVT GALELLQQLF RTPPPELLQT LTAV GGIGQ LTAAKEESGG RSRSGSIVEL IAGGGSSCSP VLSRKQKGKV LLGEEEALED DSESRSDVSS SALTASVKDE ISGEL AASS GVSTPGSAGH DIITEQPRSQ HTLQADSVDL ASCDLTSSAT DGDEEDILSH SSSQVSAVPS DPAMDLNDGT QASSPI SDS SQTTTEGPDS AVTPSDSSEI VLDGTDNQYL GLQIGQPQDE DEEATGILPD EASEAFRNSS MALQQAHLLK NMSHCRQ PS DSSVDKFVLR DEATEPGDQE NKPCRIKGDI GQSTDDDSAP LVHCVRLLSA SFLLTGGKNV LVPDRDVRVS VKALALSC V GAAVALHPES FFSKLYKVPL DTTEYPEEQY VSDILNYIDH GDPQVRGATA ILCGTLICSI LSRSRFHVGD WMGTIRTLT GNTFSLADCI PLLRKTLKDE SSVTCKLACT AVRNCVMSLC SSSYSELGLQ LIIDVLTLRN SSYWLVRTEL LETLAEIDFR LVSFLEAKA ENLHRGAHHY TGLLKLQERV LNNVVIHLLG DEDPRVRHVA AASLIRLVPK LFYKCDQGQA DPVVAVARDQ S SVYLKLLM HETQPPSHFS VSTITRIYRG YNLLPSITDV TMENNLSRVI AAVSHELITS TTRALTFGCC EALCLLSTAF PV CIWSLGW HCGVPPLSAS DESRKSCTVG MATMILTLLS SAWFPLDLSA HQDALILAGN LLAASAPKSL RSSWASEEEA NPA ATKQEE VWPALGDRAL VPMVEQLFSH LLKVINICAH VLDDVAPGPA IKAALPSLTN PPSLSPIRRK GKEKEPGEQA SVPL SPKKG SEASAASRQS DTSGPVTTSK SSSLGSFYHL PSYLKLHDVL KATHANYKVT LDLQNSTEKF GGFLRSALDV LSQIL ELAT LQDIGKCVEE ILGYLKSCFS REPMMATVCV QQLLKTLFGT NLASQFDGLS SNPSKSQGRA QRLGSSSVRP GLYHYC FMA PYTHFTQALA DASLRNMVQA EQENDTSGWF DVLQKVSTQL KTNLTSVTKN RADKNAIHNH IRLFEPLVIK ALKQYTT TT CVQLQKQVLD LLAQLVQLRV NYCLLDSDQV FIGFVLKQFE YIEVGQFRES EAIIPNIFFF LVLLSYERYH SKQIIGIP K IIQLCDGIMA SGRKAVTHAI PALQPIVHDL FVLRGTNKAD AGKELETQKE VVVSMLLRLI QYHQVLEMFI LVLQQCHKE NEDKWKRLSR QIADIILPML AKQQMHIDSH EALGVLNTLF EILAPSSLRP VDMLLRSMFV TPNTMASVST VQLWISGILA ILRVLISQS TEDIVLSRIQ ELSFSPYLIS CTVINRLRDG DSTSTLEEHS EGKQIKNLPE ETFSRFLLQL VGILLEDIVT K QLKVEMSE QQHTFYCQEL GTLLMCLIHI FKSGMFRRIT AAATRLFRSD GCGGSFYTLD SLNLRARSMI TTHPALVLLW CQ ILLLVNH TDYRWWAEVQ QTPKRHSLSS TKLLSPQMSG EEEDSDLAAK LGMCNREIVR RGALILFCDY VCQNLHDSEH LTW LIVNHI QDLISLSHEP PVQDFISAVH RNSAASGLFI QAIQSRCENL STPTMLKKTL QCLEGIHLSQ SGAVLTLYVD RLLC TPFRV LARMVDILAC RRVEMLLAAN LQSSMAQLPM EELNRIQEYL QSSGLAQRHQ RLYSLLDRFR LSTMQDSLSP SPPVS SHPL DGDGHVSLET VSPDKDWYVH LVKSQCWTRS DSALLEGAEL VNRIPAEDMN AFMMNSEFNL SLLAPCLSLG MSEISG GQK SALFEAAREV TLARVSGTVQ QLPAVHHVFQ PELPAEPAAY WSKLNDLFGD AALYQSLPTL ARALAQYLVV VSKLPSH LH LPPEKEKDIV KFVVATLEAL SWHLIHEQIP LSLDLQAGLD CCCLALQLPG LWSVVSSTEF VTHACSLIHC VHFILEAV A VQPGEQLLSP ERRTNTPKAI SEEEEEVDPN TQNPKYITAA CEMVAEMVES LQSVLALGHK RNSGVPAFLT PLLRNIIIS LARLPLVNSY TRVPPLVWKL GWSPKPGGDF GTAFPEIPVE FLQEKEVFKE FIYRINTLGW TSRTQFEETW ATLLGVLVTQ PLVMEQEES PPEEDTERTQ INVLAVQAIT SLVLSAMTVP VAGNPAVSCL EQQPRNKPLK ALDTRFGRKL SIIRGIVEQE I QAMVSKRE NIATHHLYQA WDPVPSLSPA TTGALISHEK LLLQINPERE LGSMSYKLGQ VSIHSVWLGN SITPLREEEW DE EEEEEAD APAPSSPPTS PVNSRKHRAG VDIHSCSQFL LELYSRWILP SSSARRTPAI LISEVVRSLL VVSDLFTERN QFE LMYVTL TELRRVHPSE DEILAQYLVP ATCKAAAVLG MDKAVAEPVS RLLESTLRSS HLPSRVGALH GILYVLECDL LDDT AKQLI PVISDYLLSN LKGIAHCVNI HSQQHVLVMC ATAFYLIENY PLDVGPEFSA SIIQMCGVML SGSEESTPSI IYHCA LRGL ERLLLSEQLS RLDAESLVKL SVDRVNVHSP HRAMAALGLM LTCMYTGKEK VSPGRTSDPN PAAPDSESVI VAMERV SVL FDRIRKGFPC EARVVARILP QFLDDFFPPQ DIMNKVIGEF LSNQQPYPQF MATVVYKVFQ TLHSTGQSSM VRDWVML SL SNFTQRAPVA MATWSLSCFF VSASTSPWVA AILPHVISRM GKLEQVDVNL FCLVATDFYR HQIEEELDRR AFQSVLEV V AAPGSPYHRL LTCLRNVHKV TTCGGSGDYK DDDDK UniProtKB: Huntingtin |
-Macromolecule #2: 40-kDa huntingtin-associated protein
| Macromolecule | Name: 40-kDa huntingtin-associated protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.342254 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHSSG RENLYFQGMA AAAAGLGGGG AGPGPEAGDF LARYRLVSNK LKKRFLRKPN VAEAGEQFGQ LGRELRAQEC LPYAAWCQL AVARCQQALF HGPGEALALT EAARLFLRQE RDARQRLVCP AAYGEPLQAA ASALGAAVRL HLELGQPAAA A ALCLELAA ...String: MHHHHHHSSG RENLYFQGMA AAAAGLGGGG AGPGPEAGDF LARYRLVSNK LKKRFLRKPN VAEAGEQFGQ LGRELRAQEC LPYAAWCQL AVARCQQALF HGPGEALALT EAARLFLRQE RDARQRLVCP AAYGEPLQAA ASALGAAVRL HLELGQPAAA A ALCLELAA ALRDLGQPAA AAGHFQRAAQ LQLPQLPLAA LQALGEAASC QLLARDYTGA LAVFTRMQRL AREHGSHPVQ SL PPPPPPA PQPGPGATPA LPAALLPPNS GSAAPSPAAL GAFSDVLVRC EVSRVLLLLL LQPPPAKLLP EHAQTLEKYS WEA FDSHGQ ESSGQLPEEL FLLLQSLVMA THEKDTEAIK SLQVEMWPLL TAEQNHLLHL VLQETISPSG QGV UniProtKB: 40-kDa huntingtin-associated protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/4 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.35 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average exposure time: 8.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)