+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21982 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PARP2/HPF1_Nucleosome complex | |||||||||
 Map data Map data | Substrate class of PARP2/HPF1 from PARP2/HPF1_Nucleosome complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | |||||||||
 Authors Authors | Halic M / Bilokapic S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Bridging of DNA breaks activates PARP2-HPF1 to modify chromatin. Authors: Silvija Bilokapic / Marcin J Suskiewicz / Ivan Ahel / Mario Halic /   Abstract: Breaks in DNA strands recruit the protein PARP1 and its paralogue PARP2 to modify histones and other substrates through the addition of mono- and poly(ADP-ribose) (PAR). In the DNA damage responses, ...Breaks in DNA strands recruit the protein PARP1 and its paralogue PARP2 to modify histones and other substrates through the addition of mono- and poly(ADP-ribose) (PAR). In the DNA damage responses, this post-translational modification occurs predominantly on serine residues and requires HPF1, an accessory factor that switches the amino acid specificity of PARP1 and PARP2 from aspartate or glutamate to serine. Poly(ADP) ribosylation (PARylation) is important for subsequent chromatin decompaction and provides an anchor for the recruitment of downstream signalling and repair factors to the sites of DNA breaks. Here, to understand the molecular mechanism by which PARP enzymes recognize DNA breaks within chromatin, we determined the cryo-electron-microscopic structure of human PARP2-HPF1 bound to a nucleosome. This showed that PARP2-HPF1 bridges two nucleosomes, with the broken DNA aligned in a position suitable for ligation, revealing the initial step in the repair of double-strand DNA breaks. The bridging induces structural changes in PARP2 that signal the recognition of a DNA break to the catalytic domain, which licenses HPF1 binding and PARP2 activation. Our data suggest that active PARP2 cycles through different conformational states to exchange NAD and substrate, which may enable PARP enzymes to act processively while bound to chromatin. The processes of PARP activation and the PARP catalytic cycle we describe can explain mechanisms of resistance to PARP inhibitors and will aid the development of better inhibitors as cancer treatments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21982.map.gz emd_21982.map.gz | 48.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21982-v30.xml emd-21982-v30.xml emd-21982.xml emd-21982.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21982.png emd_21982.png | 94.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21982 http://ftp.pdbj.org/pub/emdb/structures/EMD-21982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21982 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21982.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21982.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Substrate class of PARP2/HPF1 from PARP2/HPF1_Nucleosome complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
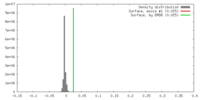
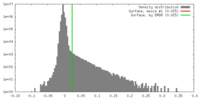
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Histone H3.2, Histone H4, Histone H2A type 1, Histone H2B 1.1, Hi...
| Entire | Name: Histone H3.2, Histone H4, Histone H2A type 1, Histone H2B 1.1, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 2/DNA Complex |
|---|---|
| Components |
|
-Supramolecule #1: Histone H3.2, Histone H4, Histone H2A type 1, Histone H2B 1.1, Hi...
| Supramolecule | Name: Histone H3.2, Histone H4, Histone H2A type 1, Histone H2B 1.1, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 2/DNA Complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 / Details: PARP2/HPF1 from PARP2/HPF1_Nucleosome complex |
|---|---|
| Molecular weight | Theoretical: 100 KDa |
-Supramolecule #2: Histone H3.2, Histone H4, Histone H2A type 1, Histone H2B 1.1
| Supramolecule | Name: Histone H3.2, Histone H4, Histone H2A type 1, Histone H2B 1.1 type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5-#6 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Supramolecule #4: Histone PARylation factor 1, Poly [ADP-ribose] polymerase 2
| Supramolecule | Name: Histone PARylation factor 1, Poly [ADP-ribose] polymerase 2 type: complex / ID: 4 / Parent: 1 / Macromolecule list: #7-#8 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 934000 |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 11000 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)