[English] 日本語
 Yorodumi
Yorodumi- EMDB-21561: The structure of human CST reveals a decameric assembly bound to ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21561 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of human CST reveals a decameric assembly bound to telomeric DNA | |||||||||||||||||||||
 Map data Map data | CST decamer sharpened map with D5 symmetry refinement | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
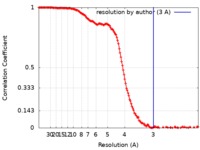
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||||||||
 Authors Authors | Lim C / Barbour AT / Zaug AJ / Goodrich KJ / McKay AE / Wuttke DS / Cech TR | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: The structure of human CST reveals a decameric assembly bound to telomeric DNA. Authors: Ci Ji Lim / Alexandra T Barbour / Arthur J Zaug / Karen J Goodrich / Allison E McKay / Deborah S Wuttke / Thomas R Cech /  Abstract: The CTC1-STN1-TEN1 (CST) complex is essential for telomere maintenance and resolution of stalled replication forks genome-wide. Here, we report the 3.0-angstrom cryo-electron microscopy structure of ...The CTC1-STN1-TEN1 (CST) complex is essential for telomere maintenance and resolution of stalled replication forks genome-wide. Here, we report the 3.0-angstrom cryo-electron microscopy structure of human CST bound to telomeric single-stranded DNA (ssDNA), which assembles as a decameric supercomplex. The atomic model of the 134-kilodalton CTC1 subunit, built almost entirely de novo, reveals the overall architecture of CST and the DNA-binding anchor site. The carboxyl-terminal domain of STN1 interacts with CTC1 at two separate docking sites, allowing allosteric mediation of CST decamer assembly. Furthermore, ssDNA appears to staple two monomers to nucleate decamer assembly. CTC1 has stronger structural similarity to Replication Protein A than the expected similarity to yeast Cdc13. The decameric structure suggests that CST can organize ssDNA analogously to the nucleosome's organization of double-stranded DNA. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21561.map.gz emd_21561.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21561-v30.xml emd-21561-v30.xml emd-21561.xml emd-21561.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21561_fsc.xml emd_21561_fsc.xml | 18.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_21561.png emd_21561.png | 88.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21561 http://ftp.pdbj.org/pub/emdb/structures/EMD-21561 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21561 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21561 | HTTPS FTP |
-Validation report
| Summary document |  emd_21561_validation.pdf.gz emd_21561_validation.pdf.gz | 78.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21561_full_validation.pdf.gz emd_21561_full_validation.pdf.gz | 78 KB | Display | |
| Data in XML |  emd_21561_validation.xml.gz emd_21561_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21561 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21561 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21561 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21561 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21561.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21561.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CST decamer sharpened map with D5 symmetry refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.078 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Decameric assembly of human CST complex with telomeric single-str...
| Entire | Name: Decameric assembly of human CST complex with telomeric single-stranded DNA |
|---|---|
| Components |
|
-Supramolecule #1: Decameric assembly of human CST complex with telomeric single-str...
| Supramolecule | Name: Decameric assembly of human CST complex with telomeric single-stranded DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Experimental: 2 MDa |
-Macromolecule #1: Homo sapiens CST complex subunit CTC1
| Macromolecule | Name: Homo sapiens CST complex subunit CTC1 / type: protein_or_peptide / ID: 1 / Details: 2XFLAG-CTC1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDYKDDDDKD YKDDDDKAAG RAQVPSSEQA WLEDAQVFIQ KTLCPAVKEP NVQLTPLVID CVKTVWLSQG RNQGSTLPLS YSFVSVQDLK THQRLPCCSH LSWSSSAYQA WAQEAGPNGN PLPREQLLLL GTLTDLSADL EQECRNGSLY VRDNTGVLSC ELIDLDLSWL ...String: MDYKDDDDKD YKDDDDKAAG RAQVPSSEQA WLEDAQVFIQ KTLCPAVKEP NVQLTPLVID CVKTVWLSQG RNQGSTLPLS YSFVSVQDLK THQRLPCCSH LSWSSSAYQA WAQEAGPNGN PLPREQLLLL GTLTDLSADL EQECRNGSLY VRDNTGVLSC ELIDLDLSWL GHLFLFPRWS YLPPARWNSS GEGHLELWDA PVPVFPLTIS PGPVTPIPVL YPESASCLLR LRNKLRGVQR NLAGSLVRLS ALVKSKQKAY FILSLGRSHP AVTHVSIIVQ VPAQLVWHRA LRPGTAYVLT ELRVSKIRGQ RQHVWMTSQS SRLLLLKPEC VQELELELEG PLLEADPKPL PMPSNSEDKK DPESLVRYSR LLSYSGAVTG VLNEPAGLYE LDGQLGLCLA YQQFRGLRRV MRPGVCLQLQ DVHLLQSVGG GTRRPVLAPC LRGAVLLQSF SRQKPGAHSS RQAYGASLYE QLVWERQLGL PLYLWATKAL EELACKLCPH VLRHHQFLQH SSPGSPSLGL QLLAPTLDLL APPGSPVRNA HNEILEEPHH CPLQKYTRLQ TPSSFPTLAT LKEEGQRKAW ASFDPKALLP LPEASYLPSC QLNRRLAWSW LCLLPSAFCP AQVLLGVLVA SSHKGCLQLR DQSGSLPCLL LAKHSQPLSD PRLIGCLVRA ERFQLIVERD VRSSFPSWKE LSMPGFIQKQ QARVYVQFFL ADALILPVPR PCLHSATPST PQTDPTGPEG PHLGQSRLFL LCHKEALMKR NFCVPPGASP EVPKPALSFY VLGSWLGGTQ RKEGTGWGLP EPQGNDDNDQ KVHLIFFGSS VRWFEFLHPG QVYRLVAPGP ATPMLFEKDG SSCISRRPLE LAGCASCLTV QDNWTLELES SQDIQDVLDA NKSLPESSLT DLLSDNFTDS LVSFSAEILS RTLCEPLVAS LWMKLGNTGA MRRCVKLTVA LETAECEFPP HLDVYIEDPH LPPSLGLLPG ARVHFSQLEK RVSRSHNVYC CFRSSTYVQV LSFPPETTIS VPLPHIYLAE LLQGGQSPFQ ATASCHIVSV FSLQLFWVCA YCTSICRQGK CTRLGSTCPT QTAISQAIIR LLVEDGTAEA VVTCRNHHVA AALGLCPREW ASLLDFVQVP GRVVLQFAGP GAQLESSARV DEPMTMFLWT LCTSPSVLRP IVLSFELERK PSKIVPLEPP RLQRFQCGEL PFLTHVNPRL RLSCLSIRES EYSSSLGILA SSC |
-Macromolecule #2: Homo sapiens CST complex subunit STN1
| Macromolecule | Name: Homo sapiens CST complex subunit STN1 / type: protein_or_peptide / ID: 2 / Details: 6xHIS-STN1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHQPG SSRCEEETPS LLWGLDPVFL AFAKLYIRDI LDMKESRQVP GVFLYNGHPI KQVDVLGTVI GVRERDAFYS YGVDDSTGVI NCICWKKLNT ESVSAAPSAA RELSLTSQLK KLQETIEQKT KIEIGDTIRV RGSIRTYREE REIHATTYYK VDDPVWNIQI ...String: MHHHHHHQPG SSRCEEETPS LLWGLDPVFL AFAKLYIRDI LDMKESRQVP GVFLYNGHPI KQVDVLGTVI GVRERDAFYS YGVDDSTGVI NCICWKKLNT ESVSAAPSAA RELSLTSQLK KLQETIEQKT KIEIGDTIRV RGSIRTYREE REIHATTYYK VDDPVWNIQI ARMLELPTIY RKVYDQPFHS SALEKEEALS NPGALDLPSL TSLLSEKAKE FLMENRVQSF YQQELEMVES LLSLANQPVI HSASSDQVNF KKDTTSKAIH SIFKNAIQLL QEKGLVFQKD DGFDNLYYVT REDKDLHRKI HRIIQQDCQK PNHMEKGCHF LHILACARLS IRPGLSEAVL QQVLELLEDQ SDIVSTMEHY YTAF |
-Macromolecule #3: Homo sapiens CST complex subunit TEN1
| Macromolecule | Name: Homo sapiens CST complex subunit TEN1 / type: protein_or_peptide / ID: 3 / Details: 6xHIS-TEN1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMS QLVVDICKLN QVLLNLNERS KEIFTNSSRR FRVVAQLLDI QPYDDEENEG FLCKLIVGNL PDFHAQILDD VAHFTQVKNV RLELYVSESL YRSTFDINCS NRTPELYDAV DLTVAIWNNG FINIKCEVID IEILNLEEVN ...String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMS QLVVDICKLN QVLLNLNERS KEIFTNSSRR FRVVAQLLDI QPYDDEENEG FLCKLIVGNL PDFHAQILDD VAHFTQVKNV RLELYVSESL YRSTFDINCS NRTPELYDAV DLTVAIWNNG FINIKCEVID IEILNLEEVN QLREFIASPM GQEFLQLSNS TGDYSQPNT |
-Macromolecule #4: 3xTEL
| Macromolecule | Name: 3xTEL / type: dna / ID: 4 Details: Three repeats of TTAGGG single-stranded DNA molecule Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TTAGGGTTAG GGTTAGGG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM HEPES-NaOH pH 7.5, 500 mM NaCl, 15 mM imidazole, 1 mM TCEP |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277.15 K / Instrument: LEICA EM GP |
| Details | 3xTELssDNA molecules were added at 1.2-fold excess of CST monomers and incubated on ice for 2 hr before applying to EM grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 28053 / Average electron dose: 60.0 e/Å2 Details: Two datasets of 0 deg (20,826 movies) and 30 deg (7,227 movies) stage-tilt were collected on the FEI Titan Krios. Collection was done with super-resolution CDS mode at 0.539 angstroms/pixel |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)