[English] 日本語
 Yorodumi
Yorodumi- EMDB-21559: Single particle cryoEM structure of V. cholerae Type IV competenc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21559 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryoEM structure of V. cholerae Type IV competence pilus secretin PilQ | |||||||||
 Map data Map data | Type IV competence pilus secretin PilQ Relion PostProcess map (postprocess.mrc) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | secretion system / outer membrane protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
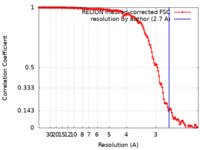
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Weaver SJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: CryoEM structure of the type IVa pilus secretin required for natural competence in Vibrio cholerae. Authors: Sara J Weaver / Davi R Ortega / Matthew H Sazinsky / Triana N Dalia / Ankur B Dalia / Grant J Jensen /  Abstract: Natural transformation is the process by which bacteria take up genetic material from their environment and integrate it into their genome by homologous recombination. It represents one mode of ...Natural transformation is the process by which bacteria take up genetic material from their environment and integrate it into their genome by homologous recombination. It represents one mode of horizontal gene transfer and contributes to the spread of traits like antibiotic resistance. In Vibrio cholerae, a type IVa pilus (T4aP) is thought to facilitate natural transformation by extending from the cell surface, binding to exogenous DNA, and retracting to thread this DNA through the outer membrane secretin, PilQ. Here, we use a functional tagged allele of VcPilQ purified from native V. cholerae cells to determine the cryoEM structure of the VcPilQ secretin in amphipol to ~2.7 Å. We use bioinformatics to examine the domain architecture and gene neighborhood of T4aP secretins in Proteobacteria in comparison with VcPilQ. This structure highlights differences in the architecture of the T4aP secretin from the type II and type III secretion system secretins. Based on our cryoEM structure, we design a series of mutants to reversibly regulate VcPilQ gate dynamics. These experiments support the idea of VcPilQ as a potential druggable target and provide insight into the channel that DNA likely traverses to promote the spread of antibiotic resistance via horizontal gene transfer by natural transformation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21559.map.gz emd_21559.map.gz | 228.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21559-v30.xml emd-21559-v30.xml emd-21559.xml emd-21559.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21559_fsc.xml emd_21559_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_21559.png emd_21559.png | 226.3 KB | ||
| Masks |  emd_21559_msk_1.map emd_21559_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21559.cif.gz emd-21559.cif.gz | 6.4 KB | ||
| Others |  emd_21559_additional_1.map.gz emd_21559_additional_1.map.gz emd_21559_half_map_1.map.gz emd_21559_half_map_1.map.gz emd_21559_half_map_2.map.gz emd_21559_half_map_2.map.gz | 192 MB 192.5 MB 192.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21559 http://ftp.pdbj.org/pub/emdb/structures/EMD-21559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21559 | HTTPS FTP |
-Related structure data
| Related structure data |  6w6mMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21559.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21559.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Type IV competence pilus secretin PilQ Relion PostProcess map (postprocess.mrc) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.104 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
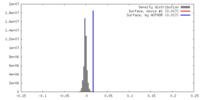
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21559_msk_1.map emd_21559_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
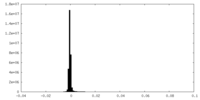
| Density Histograms |
-Additional map: Type IV competence pilus secretin PilQ Relion Refine3D...
| File | emd_21559_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Type IV competence pilus secretin PilQ Relion Refine3D output map (run_class001.mrc) | ||||||||||||
| Projections & Slices |
| ||||||||||||
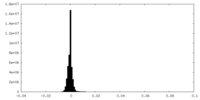
| Density Histograms |
-Half map: Type IV competence pilus secretin PilQ Relion Refine3D half map 1
| File | emd_21559_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Type IV competence pilus secretin PilQ Relion Refine3D half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Type IV competence pilus secretin PilQ Relion Refine3D half map 2
| File | emd_21559_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Type IV competence pilus secretin PilQ Relion Refine3D half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
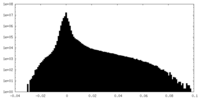
| Density Histograms |
- Sample components
Sample components
-Entire : Vibrio cholerae Type IV competence pilus secretin PilQ
| Entire | Name: Vibrio cholerae Type IV competence pilus secretin PilQ |
|---|---|
| Components |
|
-Supramolecule #1: Vibrio cholerae Type IV competence pilus secretin PilQ
| Supramolecule | Name: Vibrio cholerae Type IV competence pilus secretin PilQ type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 826 KDa |
-Macromolecule #1: Type IV pilus secretin PilQ family protein
| Macromolecule | Name: Type IV pilus secretin PilQ family protein / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.459188 KDa |
| Sequence | String: MRNGLKTYVA QTWLTLWVGL ALCASSTVFS AEFATTNQLE NIDFRVNKEK AAVLIVELAS PSAVVDVQKV QEGLNIELLK TDVADDKLY LLDVKDFSTP VESVEVFRKA PSTQLVVTVD GEFQHDYTLK GKYLEVVISK LKADEKPKPK SVLEKEGKLI S INFQDIPV ...String: MRNGLKTYVA QTWLTLWVGL ALCASSTVFS AEFATTNQLE NIDFRVNKEK AAVLIVELAS PSAVVDVQKV QEGLNIELLK TDVADDKLY LLDVKDFSTP VESVEVFRKA PSTQLVVTVD GEFQHDYTLK GKYLEVVISK LKADEKPKPK SVLEKEGKLI S INFQDIPV RNVLQLIADY NGFNLVVSDS VVGNLTLRLD GVPWQQVLDI ILQVKGLDKR VDGNVILIAP KEELDLREKQ AL EKARLAE ELGDLKSEII KINFAKASDI AAMIGGEGNV NMLSERGSIS IDERTNSLLI RELPDNIAVI REIIESLDIP VKQ VQIEAR IVTVKEGNLE ELGVRWGVMS TNGSHSVGGS IESNLWQKGL LADDEFPVDE FLNVNLASTS ANASSIAFQV AKLG SGTLL DLELSALQNE SKAEIISSPR LITTNKQPAY IEQGTEIPYL ESSSSGASTV AFKKAVLSLK VTPQITPDNR LVLDL SVTQ DRRGETVKTG TGEAVSIDTQ RIGTQVLVNN GETVVLGGIF QHSINNSVDK VPLLGDLPVL GALFRRTYEQ MGKSEL LIF VTPKVVIQ UniProtKB: Type IV pilus secretin PilQ family protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: Pelco EasiGlow, 20 mA, 60 seconds | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV / Details: blot force -6, blot time 4 s. | |||||||||
| Details | The protein was purified using Ni NTA agarose beads (Anatrace, SUPER-NINTA25) and concentrated to ~1 mg/mL. PilQ was then exchanged into Amphipol A8-35 (0.585 mg for a 3:1 ratio, Anatrace, A835). BioBeads were used to remove excess DDM and the sample was concentrated to ~0.8 mg/mL |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 3808 / Average exposure time: 3.7 sec. / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 30.0 µm / Nominal defocus min: 10.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6w6m: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)