+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21390 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of nevanimibe-bound human tetrameric ACAT1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cholesterol / cholesteryl ester / acyltransferase / MBOAT / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsterol O-acyltransferase / sterol O-acyltransferase activity / cholesterol O-acyltransferase activity / macrophage derived foam cell differentiation / cholesterol storage / positive regulation of amyloid precursor protein biosynthetic process / very-low-density lipoprotein particle assembly / fatty-acyl-CoA binding / low-density lipoprotein particle clearance / LDL clearance ...sterol O-acyltransferase / sterol O-acyltransferase activity / cholesterol O-acyltransferase activity / macrophage derived foam cell differentiation / cholesterol storage / positive regulation of amyloid precursor protein biosynthetic process / very-low-density lipoprotein particle assembly / fatty-acyl-CoA binding / low-density lipoprotein particle clearance / LDL clearance / cholesterol efflux / cholesterol binding / cholesterol metabolic process / cholesterol homeostasis / endoplasmic reticulum membrane / endoplasmic reticulum / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.67 Å | |||||||||
 Authors Authors | Li X / Long T | |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structure of nevanimibe-bound tetrameric human ACAT1. Authors: Tao Long / Yingyuan Sun / Abdirahman Hassan / Xiaofeng Qi / Xiaochun Li /  Abstract: Cholesterol is an essential component of mammalian cell membranes, constituting up to 50% of plasma membrane lipids. By contrast, it accounts for only 5% of lipids in the endoplasmic reticulum (ER). ...Cholesterol is an essential component of mammalian cell membranes, constituting up to 50% of plasma membrane lipids. By contrast, it accounts for only 5% of lipids in the endoplasmic reticulum (ER). The ER enzyme sterol O-acyltransferase 1 (also named acyl-coenzyme A:cholesterol acyltransferase, ACAT1) transfers a long-chain fatty acid to cholesterol to form cholesteryl esters that coalesce into cytosolic lipid droplets. Under conditions of cholesterol overload, ACAT1 maintains the low cholesterol concentration of the ER and thereby has an essential role in cholesterol homeostasis. ACAT1 has also been implicated in Alzheimer's disease, atherosclerosis and cancers. Here we report a cryo-electron microscopy structure of human ACAT1 in complex with nevanimibe, an inhibitor that is in clinical trials for the treatment of congenital adrenal hyperplasia. The ACAT1 holoenzyme is a tetramer that consists of two homodimers. Each monomer contains nine transmembrane helices (TMs), six of which (TM4-TM9) form a cavity that accommodates nevanimibe and an endogenous acyl-coenzyme A. This cavity also contains a histidine that has previously been identified as essential for catalytic activity. Our structural data and biochemical analyses provide a physical model to explain the process of cholesterol esterification, as well as details of the interaction between nevanimibe and ACAT1, which may help to accelerate the development of ACAT1 inhibitors to treat related diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21390.map.gz emd_21390.map.gz | 11.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21390-v30.xml emd-21390-v30.xml emd-21390.xml emd-21390.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21390.png emd_21390.png | 41.4 KB | ||
| Filedesc metadata |  emd-21390.cif.gz emd-21390.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21390 http://ftp.pdbj.org/pub/emdb/structures/EMD-21390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21390 | HTTPS FTP |
-Related structure data
| Related structure data |  6vumMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21390.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21390.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ACAT1
| Entire | Name: ACAT1 |
|---|---|
| Components |
|
-Supramolecule #1: ACAT1
| Supramolecule | Name: ACAT1 / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sterol O-acyltransferase 1
| Macromolecule | Name: Sterol O-acyltransferase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: sterol O-acyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 65.80125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVGEEKMSLR NRLSKSRENP EEDEDQRNPA KESLETPSNG RIDIKQLIAK KIKLTAEAEE LKPFFMKEVG SHFDDFVTNL IEKSASLDN GGCALTTFSV LEGEKNNHRA KDLRAPPEQG KIFIARRSLL DELLEVDHIR TIYHMFIALL ILFILSTLVV D YIDEGRLV ...String: MVGEEKMSLR NRLSKSRENP EEDEDQRNPA KESLETPSNG RIDIKQLIAK KIKLTAEAEE LKPFFMKEVG SHFDDFVTNL IEKSASLDN GGCALTTFSV LEGEKNNHRA KDLRAPPEQG KIFIARRSLL DELLEVDHIR TIYHMFIALL ILFILSTLVV D YIDEGRLV LEFSLLSYAF GKFPTVVWTW WIMFLSTFSV PYFLFQHWAT GYSKSSHPLI RSLFHGFLFM IFQIGVLGFG PT YVVLAYT LPPASRFIII FEQIRFVMKA HSFVRENVPR VLNSAKEKSS TVPIPTVNQY LYFLFAPTLI YRDSYPRNPT VRW GYVAMK FAQVFGCFFY VYYIFERLCA PLFRNIKQEP FSARVLVLCV FNSILPGVLI LFLTFFAFLH CWLNAFAEML RFGD RMFYK DWWNSTSYSN YYRTWNVVVH DWLYYYAYKD FLWFFSKRFK SAAMLAVFAV SAVVHEYALA VCLSFFYPVL FVLFM FFGM AFNFIVNDSR KKPIWNVLMW TSLFLGNGVL LCFYSQEWYA RQHCPLKNPT FLDYVRPRSW TCRYVFDYKD DDDK UniProtKB: Sterol O-acyltransferase 1 |
-Macromolecule #2: OLEIC ACID
| Macromolecule | Name: OLEIC ACID / type: ligand / ID: 2 / Number of copies: 4 / Formula: OLA |
|---|---|
| Molecular weight | Theoretical: 282.461 Da |
| Chemical component information |  ChemComp-OLA: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 10 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #4: nevanimibe
| Macromolecule | Name: nevanimibe / type: ligand / ID: 4 / Number of copies: 4 / Formula: ROV |
|---|---|
| Molecular weight | Theoretical: 421.618 Da |
| Chemical component information | 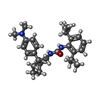 ChemComp-ROV: |
-Macromolecule #5: COENZYME A
| Macromolecule | Name: COENZYME A / type: ligand / ID: 5 / Number of copies: 2 / Formula: COA |
|---|---|
| Molecular weight | Theoretical: 767.534 Da |
| Chemical component information |  ChemComp-COA: |
-Macromolecule #6: S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydrox...
| Macromolecule | Name: S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza- ...Name: S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) type: ligand / ID: 6 / Number of copies: 2 / Formula: 3VV |
|---|---|
| Molecular weight | Theoretical: 1.03198 KDa |
| Chemical component information |  ChemComp-3VV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.67 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 263839 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















