[English] 日本語
 Yorodumi
Yorodumi- EMDB-20896: Negative-stain EM map from 3D sorting of BG505-immunized rhesus m... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20896 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
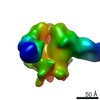
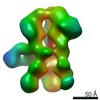
| Title | Negative-stain EM map from 3D sorting of BG505-immunized rhesus macaque 12-137 wk 28 serum fab complexed with BG505 SOSIP.664 trimer | |||||||||
 Map data Map data | Negative-stain EM map from 3D sorting of BG505-immunized rhesus macaque 12-137 wk 28 serum fab complexed with BG505 SOSIP.664 trimer | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Nogal B / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2020 Journal: Cell Rep / Year: 2020Title: Mapping Polyclonal Antibody Responses in Non-human Primates Vaccinated with HIV Env Trimer Subunit Vaccines. Authors: Bartek Nogal / Matteo Bianchi / Christopher A Cottrell / Robert N Kirchdoerfer / Leigh M Sewall / Hannah L Turner / Fangzhu Zhao / Devin Sok / Dennis R Burton / Lars Hangartner / Andrew B Ward /  Abstract: Rational immunogen design aims to focus antibody responses to vulnerable sites on primary antigens. Given the size of these antigens, there is, however, potential for eliciting unwanted, off-target ...Rational immunogen design aims to focus antibody responses to vulnerable sites on primary antigens. Given the size of these antigens, there is, however, potential for eliciting unwanted, off-target responses. Here, we use our electron microscopy polyclonal epitope mapping approach to describe the antibody specificities elicited by immunization of non-human primates with soluble HIV envelope trimers and subsequent repeated viral challenge. An increased diversity of epitopes recognized and the approach angle by which these antibodies bind constitute a hallmark of the humoral response in most protected animals. We also show that fusion peptide-specific antibodies are likely responsible for some neutralization breadth. Moreover, cryoelectron microscopy (cryo-EM) analysis of a fully protected animal reveals a high degree of clonality within a subset of putatively neutralizing antibodies, enabling a detailed molecular description of the antibody paratope. Our results provide important insights into the immune response against a vaccine candidate that entered into clinical trials in 2019. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20896.map.gz emd_20896.map.gz | 6.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20896-v30.xml emd-20896-v30.xml emd-20896.xml emd-20896.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20896.png emd_20896.png | 55 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20896 http://ftp.pdbj.org/pub/emdb/structures/EMD-20896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20896 | HTTPS FTP |
-Validation report
| Summary document |  emd_20896_validation.pdf.gz emd_20896_validation.pdf.gz | 78 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20896_full_validation.pdf.gz emd_20896_full_validation.pdf.gz | 77.1 KB | Display | |
| Data in XML |  emd_20896_validation.xml.gz emd_20896_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20896 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20896 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20896 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20896 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20896.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20896.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative-stain EM map from 3D sorting of BG505-immunized rhesus macaque 12-137 wk 28 serum fab complexed with BG505 SOSIP.664 trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Polyclonal serum fab with BG505 SOSIP.664
| Entire | Name: Polyclonal serum fab with BG505 SOSIP.664 |
|---|---|
| Components |
|
-Supramolecule #1: Polyclonal serum fab with BG505 SOSIP.664
| Supramolecule | Name: Polyclonal serum fab with BG505 SOSIP.664 / type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: BG505 SOSIPv5.2 trimer
| Supramolecule | Name: BG505 SOSIPv5.2 trimer / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism: unidentified (others) |
-Supramolecule #3: Polyclonal Fab
| Supramolecule | Name: Polyclonal Fab / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 4.5 |
|---|---|
| Staining | Type: NEGATIVE / Material: uranyl formate |
| Grid | Details: unspecified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 72000 |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 3200 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera












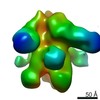

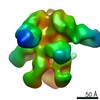

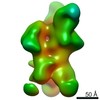
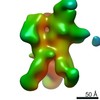
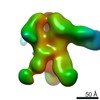
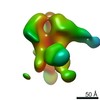
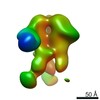

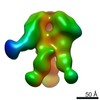

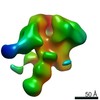


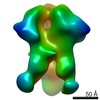
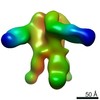

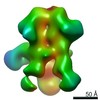
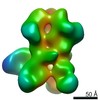

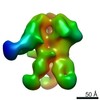




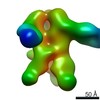
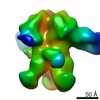
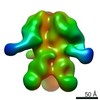

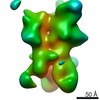
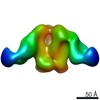


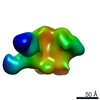

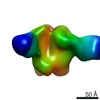
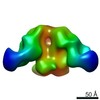


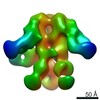




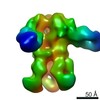








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















