[English] 日本語
 Yorodumi
Yorodumi- EMDB-20862: Extended Sensor Paddles with Bound Lipids Revealed in Mechanosens... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20862 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Extended Sensor Paddles with Bound Lipids Revealed in Mechanosensitive Channel YnaI | |||||||||
 Map data Map data | Structure of a mechanosensitive channel | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mechanosensitive channel / Phosphoserine lipid / complete paddle / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmechanosensitive monoatomic ion channel activity / cellular response to osmotic stress / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
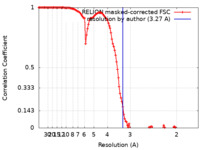
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Hu W / Wang Z | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Mechanosensitive channel YnaI has lipid-bound extended sensor paddles. Authors: Wenxin Hu / Zhiming Wang / Hongjin Zheng /  Abstract: The general mechanism of bacterial mechanosensitive channels (MS) has been characterized by extensive studies on a small conductance channel MscS from Escherichia coli (E. coli). However, recent ...The general mechanism of bacterial mechanosensitive channels (MS) has been characterized by extensive studies on a small conductance channel MscS from Escherichia coli (E. coli). However, recent structural studies on the same channel have revealed controversial roles of various channel-bound lipids in channel gating. To better understand bacterial MscS-like channels, it is necessary to characterize homologs other than MscS. Here, we describe the structure of YnaI, one of the closest MscS homologs in E. coli, in its non-conducting state at 3.3 Å resolution determined by cryo electron microscopy. Our structure revealed the intact membrane sensor paddle domain in YnaI, which was stabilized by functionally important residues H43, Q46, Y50 and K93. In the pockets between sensor paddles, there were clear lipid densities that interact strongly with residues Q100 and R120. These lipids were a mixture of natural lipids but may be enriched in cardiolipin and phosphatidylserine. In addition, residues along the ion-conducting pathway and responsible for the heptameric assembly were discussed. Together with biochemical experiments and mutagenesis studies, our results provide strong support for the idea that the pocket lipids are functionally important for mechanosensitive channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20862.map.gz emd_20862.map.gz | 130.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20862-v30.xml emd-20862-v30.xml emd-20862.xml emd-20862.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20862_fsc.xml emd_20862_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_20862.png emd_20862.png | 158.7 KB | ||
| Filedesc metadata |  emd-20862.cif.gz emd-20862.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20862 http://ftp.pdbj.org/pub/emdb/structures/EMD-20862 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20862 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20862 | HTTPS FTP |
-Related structure data
| Related structure data |  6urtMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20862.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20862.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of a mechanosensitive channel | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.864 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : YnaI with PS lipids
| Entire | Name: YnaI with PS lipids |
|---|---|
| Components |
|
-Supramolecule #1: YnaI with PS lipids
| Supramolecule | Name: YnaI with PS lipids / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 270 KDa |
-Macromolecule #1: Low conductance mechanosensitive channel YnaI
| Macromolecule | Name: Low conductance mechanosensitive channel YnaI / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.792344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIAELFTNNA LNLVIIFGSC AALILMSFWF RRGNRKRKGF LFHAVQFLIY TIIISAVGSI INYVIENYKL KFITPGVIDF ICTSLIAVI LTIKLFLLIN QFEKQQIKKG RDITSARIMS RIIKITIIVV LVLLYGEHFG MSLSGLLTFG GIGGLAVGMA G KDILSNFF ...String: MIAELFTNNA LNLVIIFGSC AALILMSFWF RRGNRKRKGF LFHAVQFLIY TIIISAVGSI INYVIENYKL KFITPGVIDF ICTSLIAVI LTIKLFLLIN QFEKQQIKKG RDITSARIMS RIIKITIIVV LVLLYGEHFG MSLSGLLTFG GIGGLAVGMA G KDILSNFF SGIMLYFDRP FSIGDWIRSP DRNIEGTVAE IGWRITKITT FDNRPLYVPN SLFSSISVEN PGRMTNRRIT TT IGLRYED AAKVGVIVEA VREMLKNHPA IDQRQTLLVY FNQFADSSLN IMVYCFTKTT VWAEWLAAQQ DVYLKIIDIV QSH GADFAF PSQTLYMDNI TPPEQGR UniProtKB: Low conductance mechanosensitive channel YnaI |
-Macromolecule #2: O-{(R)-hydroxy[(2R)-3-(icosyloxy)-2-(tetradecanoyloxy)propoxy]pho...
| Macromolecule | Name: O-{(R)-hydroxy[(2R)-3-(icosyloxy)-2-(tetradecanoyloxy)propoxy]phosphoryl}-L-serine type: ligand / ID: 2 / Number of copies: 7 / Formula: QGD |
|---|---|
| Molecular weight | Theoretical: 750.038 Da |
| Chemical component information |  ChemComp-QGD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20mM HEPES pH7.5, 150mM NaCl, 0.01% LMNG, 3mM Fluorinated Fos-Cholin 8 |
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.0 sec. / Average electron dose: 64.3 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)