[English] 日本語
 Yorodumi
Yorodumi- EMDB-20590: CryoEM reconstruction of membrane-bound ESCRT-III filament compos... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20590 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM reconstruction of membrane-bound ESCRT-III filament composed of CHMP1B only | ||||||||||||||||||
 Map data Map data | RELION Refine3D filtered map with helical symmetry imposed on the central 40% | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | membrane remodeling / membrane-bound protein filament / ESCRT-III / LIPID BINDING PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMIT domain binding / multivesicular body-lysosome fusion / amphisome membrane / vesicle fusion with vacuole / ESCRT III complex disassembly / late endosome to lysosome transport / ESCRT III complex / kinetochore microtubule / endosome transport via multivesicular body sorting pathway / membrane coat ...MIT domain binding / multivesicular body-lysosome fusion / amphisome membrane / vesicle fusion with vacuole / ESCRT III complex disassembly / late endosome to lysosome transport / ESCRT III complex / kinetochore microtubule / endosome transport via multivesicular body sorting pathway / membrane coat / nuclear membrane reassembly / multivesicular body sorting pathway / regulation of centrosome duplication / midbody abscission / membrane fission / plasma membrane repair / late endosome to vacuole transport / ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway / multivesicular body assembly / multivesicular body membrane / regulation of mitotic spindle assembly / mitotic metaphase chromosome alignment / nucleus organization / viral budding via host ESCRT complex / autophagosome membrane / autophagosome maturation / nuclear pore / multivesicular body / viral budding from plasma membrane / establishment of protein localization / kinetochore / autophagy / protein transport / midbody / endosome membrane / protein domain specific binding / cell division / lysosomal membrane / extracellular exosome / nucleoplasm / identical protein binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
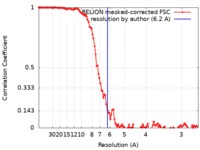
| Method | helical reconstruction / cryo EM / Resolution: 6.2 Å | ||||||||||||||||||
 Authors Authors | Nguyen HC / Frost A | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: Membrane constriction and thinning by sequential ESCRT-III polymerization. Authors: Henry C Nguyen / Nathaniel Talledge / John McCullough / Abhimanyu Sharma / Frank R Moss / Janet H Iwasa / Michael D Vershinin / Wesley I Sundquist / Adam Frost /  Abstract: The endosomal sorting complexes required for transport (ESCRTs) mediate diverse membrane remodeling events. These typically require ESCRT-III proteins to stabilize negatively curved membranes; ...The endosomal sorting complexes required for transport (ESCRTs) mediate diverse membrane remodeling events. These typically require ESCRT-III proteins to stabilize negatively curved membranes; however, recent work has indicated that certain ESCRT-IIIs also participate in positive-curvature membrane-shaping reactions. ESCRT-IIIs polymerize into membrane-binding filaments, but the structural basis for negative versus positive membrane remodeling by these proteins remains poorly understood. To learn how certain ESCRT-IIIs shape positively curved membranes, we determined structures of human membrane-bound CHMP1B-only, membrane-bound CHMP1B + IST1, and IST1-only filaments by cryo-EM. Our structures show how CHMP1B first polymerizes into a single-stranded helical filament, shaping membranes into moderate-curvature tubules. Subsequently, IST1 assembles a second strand on CHMP1B, further constricting the membrane tube and reducing its diameter nearly to the fission point. Each step of constriction thins the underlying bilayer, lowering the barrier to membrane fission. Our structures reveal how a two-component, sequential polymerization mechanism drives membrane tubulation, constriction and bilayer thinning. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20590.map.gz emd_20590.map.gz | 100.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20590-v30.xml emd-20590-v30.xml emd-20590.xml emd-20590.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20590_fsc.xml emd_20590_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_20590.png emd_20590.png | 198.5 KB | ||
| Masks |  emd_20590_msk_1.map emd_20590_msk_1.map | 129.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20590.cif.gz emd-20590.cif.gz | 6 KB | ||
| Others |  emd_20590_additional_1.map.gz emd_20590_additional_1.map.gz emd_20590_additional_2.map.gz emd_20590_additional_2.map.gz emd_20590_additional_3.map.gz emd_20590_additional_3.map.gz emd_20590_additional_4.map.gz emd_20590_additional_4.map.gz emd_20590_half_map_1.map.gz emd_20590_half_map_1.map.gz emd_20590_half_map_2.map.gz emd_20590_half_map_2.map.gz | 35.2 MB 101.9 MB 102 MB 102 MB 101.4 MB 101.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20590 http://ftp.pdbj.org/pub/emdb/structures/EMD-20590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20590 | HTTPS FTP |
-Related structure data
| Related structure data |  6tz9MC  6tz4C  6tz5C  6tzaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10397 (Title: CryoEM reconstruction of membrane-bound ESCRT-III filament composed of CHMP1B only EMPIAR-10397 (Title: CryoEM reconstruction of membrane-bound ESCRT-III filament composed of CHMP1B onlyData size: 3.8 Data #1: Processed particle stack and metadata for a cryoEM helical reconstruction of membrane-bound ESCRT-III filament composed of CHMP1B only [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20590.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20590.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D filtered map with helical symmetry imposed on the central 40% | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.345 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
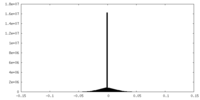
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20590_msk_1.map emd_20590_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
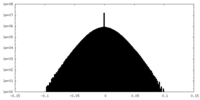
| Density Histograms |
-Additional map: RELION postprocessed map with helical symmetry applied to...
| File | emd_20590_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION postprocessed map with helical symmetry applied to the entire map | ||||||||||||
| Projections & Slices |
| ||||||||||||
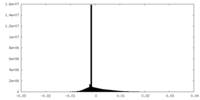
| Density Histograms |
-Additional map: RELION Refine3D filtered map with no symmetry applied (C1 map)
| File | emd_20590_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D filtered map with no symmetry applied (C1 map) | ||||||||||||
| Projections & Slices |
| ||||||||||||
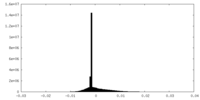
| Density Histograms |
-Additional map: RELION Refine3D unfiltered half map1 with no symmetry...
| File | emd_20590_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D unfiltered half map1 with no symmetry applied (C1 half1 map) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: RELION Refine3D unfiltered half map2 with no symmetry...
| File | emd_20590_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D unfiltered half map2 with no symmetry applied (C1 half2 map) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: RELION Refine3D unfiltered half map1 with helical symmetry...
| File | emd_20590_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D unfiltered half map1 with helical symmetry imposed on the central 40% | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: RELION Refine3D unfiltered half map2 with helical symmetry...
| File | emd_20590_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D unfiltered half map2 with helical symmetry imposed on the central 40% | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : membrane-bound ESCRT-III copolymer filament composed of CHMP1B only
| Entire | Name: membrane-bound ESCRT-III copolymer filament composed of CHMP1B only |
|---|---|
| Components |
|
-Supramolecule #1: membrane-bound ESCRT-III copolymer filament composed of CHMP1B only
| Supramolecule | Name: membrane-bound ESCRT-III copolymer filament composed of CHMP1B only type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Charged multivesicular body protein 1b
| Macromolecule | Name: Charged multivesicular body protein 1b / type: protein_or_peptide / ID: 1 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.140354 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNMEKHLFN LKFAAKELSR SAKKCDKEEK AEKAKIEKAI QKGNMEVARI HAENAIRQKN QAVNFLRMSA RVDAVAARVQ TAVTMGKVT KSMAGVVKSM DATLKTMNLE KISALMDKFE HQFETLDVQT QQMEDTMSST TTLTTPQNQV DMLLQEMADE A GLDLNMEL ...String: MSNMEKHLFN LKFAAKELSR SAKKCDKEEK AEKAKIEKAI QKGNMEVARI HAENAIRQKN QAVNFLRMSA RVDAVAARVQ TAVTMGKVT KSMAGVVKSM DATLKTMNLE KISALMDKFE HQFETLDVQT QQMEDTMSST TTLTTPQNQV DMLLQEMADE A GLDLNMEL PQGQTGSVGT SVASAEQDEL SQRLARLRDQ V UniProtKB: Charged multivesicular body protein 1b |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 292 K / Instrument: FEI VITROBOT MARK III Details: Grids were blotted with Whatman No. 1 filter paper for 4-8 seconds with a 0 mm offset at 19C and 100 percent humidity before plunging into liquid ethane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)