+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2059 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HBV T=4 pgRNA-filled Cp183 capsid | |||||||||
 Map data Map data | Cryo-EM structure of HBV T=4 pgRNA-filled Cp183 capsid (Cp183RNA-SSS) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HBV / Cp183 / pgRNA | |||||||||
| Biological species |   Hepatitis B virus Hepatitis B virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||
 Authors Authors | Wang JC-Y / Dhasan RS / Zlotnick A | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2012 Journal: PLoS Pathog / Year: 2012Title: Structural organization of pregenomic RNA and the carboxy-terminal domain of the capsid protein of hepatitis B virus. Authors: Joseph C-Y Wang / Mary S Dhason / Adam Zlotnick /  Abstract: The Hepatitis B Virus (HBV) double-stranded DNA genome is reverse transcribed from its RNA pregenome (pgRNA) within the virus core (or capsid). Phosphorylation of the arginine-rich carboxy-terminal ...The Hepatitis B Virus (HBV) double-stranded DNA genome is reverse transcribed from its RNA pregenome (pgRNA) within the virus core (or capsid). Phosphorylation of the arginine-rich carboxy-terminal domain (CTD) of the HBV capsid protein (Cp183) is essential for pgRNA encapsidation and reverse transcription. However, the structure of the CTD remains poorly defined. Here we report sub-nanometer resolution cryo-EM structures of in vitro assembled empty and pgRNA-filled Cp183 capsids in unphosphorylated and phosphorylation-mimic states. In empty capsids, we found unexpected evidence of surface accessible CTD density partially occluding pores in the capsid surface. We also observed that CTD organization changed substantively as a function of phosphorylation. In RNA-filled capsids, unphosphorylated CTDs favored thick ropes of RNA, while the phosphorylation-mimic favored a mesh of thin, high-density strands suggestive of single stranded RNA. These results demonstrate that the CTD can regulate nucleic acid structure, supporting the hypothesis that the HBV capsid has a functional role as a nucleic acid chaperone. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2059.map.gz emd_2059.map.gz | 41.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2059-v30.xml emd-2059-v30.xml emd-2059.xml emd-2059.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2059.jpg EMD-2059.jpg | 266.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2059 http://ftp.pdbj.org/pub/emdb/structures/EMD-2059 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2059 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2059 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2059.map.gz / Format: CCP4 / Size: 101.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2059.map.gz / Format: CCP4 / Size: 101.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of HBV T=4 pgRNA-filled Cp183 capsid (Cp183RNA-SSS) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4836 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
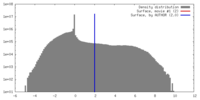
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HBV T=4 pgRNA-filled Cp183 capsid
| Entire | Name: HBV T=4 pgRNA-filled Cp183 capsid |
|---|---|
| Components |
|
-Supramolecule #1000: HBV T=4 pgRNA-filled Cp183 capsid
| Supramolecule | Name: HBV T=4 pgRNA-filled Cp183 capsid / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Hepatitis B virus
| Supramolecule | Name: Hepatitis B virus / type: virus / ID: 1 / Name.synonym: HBV Cp183 Details: Reassembled the purified Cp183 dimers with in vitro transcribed HBV pgRNA at a molar ratio of protein dimer to RNA polymer = 120:1 NCBI-ID: 10407 / Sci species name: Hepatitis B virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: Yes / Syn species name: HBV Cp183 |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Virus shell | Shell ID: 1 / Name: Cp183 / T number (triangulation number): 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 150 mM NaCl, 50 mM HEPES, 2 mM DTT |
|---|---|
| Grid | Details: Quantifoil R 2/2 holey carbon 200 mesh copper grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 93 K / Instrument: FEI VITROBOT MARK III / Method: Blot for 4 seconds before plunging |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | JEOL 3200FS |
| Temperature | Average: 97 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 80,000 times magnification |
| Specialist optics | Energy filter - Name: Omega filter |
| Date | Apr 27, 2010 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 193 / Average electron dose: 14 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.1 mm / Nominal defocus max: 3.8 µm / Nominal defocus min: 0.27 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
- Electron microscopy #2
Electron microscopy #2
| Microscopy ID | 2 |
|---|---|
| Microscope | JEOL 3200FS |
| Temperature | Average: 97 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 80,000 times magnification |
| Specialist optics | Energy filter - Name: Omega filter |
| Date | May 4, 2010 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 193 / Average electron dose: 14 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.1 mm / Nominal defocus max: 3.8 µm / Nominal defocus min: 0.27 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle phase-flipping |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 8.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Auto3dem / Number images used: 7201 |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)