[English] 日本語
 Yorodumi
Yorodumi- EMDB-20581: In situ structure of BmCPV RNA dependent RNA polymerase at quiesc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20581 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of BmCPV RNA dependent RNA polymerase at quiescent state | |||||||||||||||||||||||||||
 Map data Map data | Sub-particle reconstruction of BmCPV in the quiescent state | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | RdRp / VIRAL PROTEIN / TRANSFERASE | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral genome replication / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / RNA binding Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |   Bombyx mori cytoplasmic polyhedrosis virus Bombyx mori cytoplasmic polyhedrosis virus | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||||||||||||||
 Authors Authors | Cui YX / Zhang YN | |||||||||||||||||||||||||||
| Funding support |  China, China,  United States, 8 items United States, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Conservative transcription in three steps visualized in a double-stranded RNA virus. Authors: Yanxiang Cui / Yinong Zhang / Kang Zhou / Jingchen Sun / Z Hong Zhou /   Abstract: Endogenous RNA transcription characterizes double-stranded RNA (dsRNA) viruses in the Reoviridae, a family that is exemplified by its simple, single-shelled member cytoplasmic polyhedrosis virus (CPV) ...Endogenous RNA transcription characterizes double-stranded RNA (dsRNA) viruses in the Reoviridae, a family that is exemplified by its simple, single-shelled member cytoplasmic polyhedrosis virus (CPV). Because of the lack of in situ structures of the intermediate stages of RNA-dependent RNA polymerase (RdRp) during transcription, it is poorly understood how RdRp detects environmental cues and internal transcriptional states to initiate and coordinate repeated cycles of transcript production inside the capsid. Here, we captured five high-resolution (2.8-3.5 Å) RdRp-RNA in situ structures-representing quiescent, initiation, early elongation, elongation and abortive states-under seven experimental conditions of CPV. We observed the 'Y'-form initial RNA fork in the initiation state and the complete transcription bubble in the elongation state. These structures reveal that de novo RNA transcription involves three major conformational changes during state transitions. Our results support an ouroboros model for endogenous conservative transcription in dsRNA viruses. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20581.map.gz emd_20581.map.gz | 58.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20581-v30.xml emd-20581-v30.xml emd-20581.xml emd-20581.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20581.png emd_20581.png | 104.6 KB | ||
| Filedesc metadata |  emd-20581.cif.gz emd-20581.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20581 http://ftp.pdbj.org/pub/emdb/structures/EMD-20581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20581 | HTTPS FTP |
-Related structure data
| Related structure data |  6ty8MC  6ty9C  6tz0C  6tz1C  6tz2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20581.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20581.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-particle reconstruction of BmCPV in the quiescent state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.062 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bombyx mori cytoplasmic polyhedrosis virus
| Entire | Name:   Bombyx mori cytoplasmic polyhedrosis virus Bombyx mori cytoplasmic polyhedrosis virus |
|---|---|
| Components |
|
-Supramolecule #1: Bombyx mori cytoplasmic polyhedrosis virus
| Supramolecule | Name: Bombyx mori cytoplasmic polyhedrosis virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / NCBI-ID: 110829 Sci species name: Bombyx mori cytoplasmic polyhedrosis virus Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: RNA-dependent RNA Polymerase
| Macromolecule | Name: RNA-dependent RNA Polymerase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bombyx mori cytoplasmic polyhedrosis virus Bombyx mori cytoplasmic polyhedrosis virus |
| Molecular weight | Theoretical: 139.007125 KDa |
| Sequence | String: MLPNTELYNT IFSETRKFTR ESFKEIEHLT AKLANDRVAR HDFLFNNSIA LISDYSGEDS NGNQLQATVT IPNEITNPKE YDPSDYPLA EDESFFKQGH KYDYLVTFRA GSLTNTYEPK TKMYKLHAAL DKLMHVKQRK SRFADLWREL CAVIASLDVW Y QTTNYPLR ...String: MLPNTELYNT IFSETRKFTR ESFKEIEHLT AKLANDRVAR HDFLFNNSIA LISDYSGEDS NGNQLQATVT IPNEITNPKE YDPSDYPLA EDESFFKQGH KYDYLVTFRA GSLTNTYEPK TKMYKLHAAL DKLMHVKQRK SRFADLWREL CAVIASLDVW Y QTTNYPLR TYVKLLFHKG DEFPFYESPS QDKIIFNDKS VASILPTFVY TCCQVGTAIM SGILTHVESI VAMNHFLHCA KD SYIDEKL KIKGIGRSWY QEALHNVGRA TVPVWSQFNE VIGHRTKTTS EPHFVSSTFI SLRAKRAELL YPEFNEYINR ALR LSKTQN DVANYYAACR AMTNDGTFLA TLTELSLDAA VFPRIEQRLV TRPAVLMSNT RHESLKQKYA NGVGSIAQSY LSSF TDEIA KRVNGIHHDE AWLNFLTTSS PGRKLTEIEK LEVGGDVAAW SNSRIVMQAV FAREYRTPER IFKSLKAPIK LVERQ QSDR RQRAISGLDN DRLFLSFMPY TIGKQIYDLN DNAAQGKQAG NAFDIGEMLY WTSQRNVLLS SIDVAGMDAS VTTNTK DIY NTFVLDVASK CTVPRFGPYY AKNMEVFEVG KRQSQVKYVN AAWQACALEA ANSQTSTSYE SEIFGQVKNA EGTYPSG RA DTSTHHTVLL QGLVRGNELK RASDGKNSCL TTIKILGDDI MEIFQGNEND THDHAVSNAS ILNESGFATT AELSQNSI V LLQQLVVNGT FWGFADRISL WTREDTKDIG RLNLAMMELN ALIDDLLFRV RRPEGLKMLG FFCGAICLRR FTLSVDNKL YDSTYNNLSK YMTLVKYDKN PDFDSTLMSL ILPLAWLFMP RGGEYPAYPF ERRDGTFTED ESMFTARGAY KRRLLYDVSN IREMIQQNS MVLDDDLLHE YGFTGALLLI DLNILDLIDE VKKEDISPVK VNELATSLEQ LGKLGEREKS RRAASDLKIR G HALSNDIV YGYGLQEKIQ KSAMATKETT VQSKRVSSRL HEVIVAKTRD YKIPTMPADA LHLYEFEVED VTVDLLPHAK HT SYSNLAY NMSFGSDGWF AFALLGGLDR SANLLRLDVA SIRGNYHKFS YDDPVFKQGY KIYKSDATLL NDFFVAISAG PKE QGILLR AFAYYSLYGN VEYHYVLSPR QLFFLSDNPV SAERLVRIPP SYYVSTQCRA LYNIFSYLHI LRSITSNQGK RLGM VLHPG LIAYVRGTSQ GAILPEADNV UniProtKB: RNA-directed RNA polymerase |
-Macromolecule #2: Viral structural protein 4
| Macromolecule | Name: Viral structural protein 4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bombyx mori cytoplasmic polyhedrosis virus Bombyx mori cytoplasmic polyhedrosis virus |
| Molecular weight | Theoretical: 63.683738 KDa |
| Sequence | String: MFAIDPLKHS KLYEEYGLYL RPHQINQEIK PTTIKKKELA PTIRSIKYAS LIHSMLAKHA ARHNGTLINP RMYADMITLG NTKVTVTKG TPKAQIDTLK MNGLTVVSKS RRNNKKKPVS DTTATIDENT DDIVTYKALT EMSTLIESFR LPSGLALIIF D DEKYQSLI ...String: MFAIDPLKHS KLYEEYGLYL RPHQINQEIK PTTIKKKELA PTIRSIKYAS LIHSMLAKHA ARHNGTLINP RMYADMITLG NTKVTVTKG TPKAQIDTLK MNGLTVVSKS RRNNKKKPVS DTTATIDENT DDIVTYKALT EMSTLIESFR LPSGLALIIF D DEKYQSLI PNYINQLIAY TQPHIIPTWQ GIADFSDTYL RSYFKRPFEL TASNLAAPQK YNLSPMTRSI FNNTGREDAV IR KLYGYGE YVFIRYEGCL ITWTGIYGEV TMMVNLSKRD LGLDVGDDYL KEYKKLLFYG VITDAIPSGI SARSTIMKIS PHK MMNPSG GALAVLSKFL EAVVSTNVIN ATLVVYAEKG AGKTSFLSTY AEQLSLASGQ VVGHLSSDAY GRWLAKNKDV EEPS FAYDY VLSLDTDDNE SYYEQKASEL LISHGISEVA QYELLSVRKK IKMMDEMNEV LIAQLENADT HSERNFYYMV STGKT TPRT LIVEGHFNAQ DATIARTDTT VLLRTINDTT QAMRDRQRGG VVQLFLRDTY YRLLPALHTT VYPFEMLESI RRWKWV H UniProtKB: Viral structural protein 4 |
-Macromolecule #3: P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE
| Macromolecule | Name: P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GTA |
|---|---|
| Molecular weight | Theoretical: 787.441 Da |
| Chemical component information | 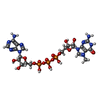 ChemComp-GTA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















