[English] 日本語
 Yorodumi
Yorodumi- EMDB-20511: Structure of full-length, fully glycosylated, non-modified HIV-1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20511 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab | |||||||||
 Map data Map data | Full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / ENV / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
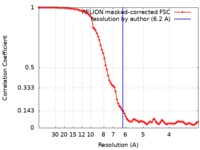
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||
 Authors Authors | Pan J / Chen B | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2020 Journal: J Mol Biol / Year: 2020Title: Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16. Authors: Junhua Pan / Hanqin Peng / Bing Chen / Stephen C Harrison /  Abstract: The HIV-1 envelope protein (Env) is the target of neutralizing antibodies and the template for vaccine immunogen design. The dynamic conformational equilibrium of trimeric Env influences its ...The HIV-1 envelope protein (Env) is the target of neutralizing antibodies and the template for vaccine immunogen design. The dynamic conformational equilibrium of trimeric Env influences its antigenicity and potential immunogenicity. Antibodies that bind at the trimer apex stabilize a "closed" conformation characteristic of the most difficult to neutralize isolates. A goal of vaccine development is therefore to mimic the closed conformation in a designed immunogen. A disulfide-stabilized, trimeric Env ectodomain-the "SOSIP" construct-has many of the relevant properties; it is also particularly suitable for structure determination. Some single-molecule studies have, however, suggested that the SOSIP trimer is not a good representation of Env on the surface of a virion or an infected cell. We isolated Env (fully cleaved to gp120 and gp41) from the surface of expressing cells using tagged, apex-binding Fab PG16 and determined the structure of the PG16-Env complex by cryo-EM to an overall resolution of 4.6 Å. Placing the only purification tag on the Fab ensured that the isolated Env was continuously stabilized in its closed, native conformation. The Env structure in this complex corresponds closely to the SOSIP structures determined by both x-ray crystallography and cryo-EM. Although the membrane-interacting elements are not resolved in our reconstruction, we can make inferences about the connection between ectodomain and membrane-proximal external region (MPER) by reference to the published cryo-tomography structure of an Env "spike" and the NMR structure of the MPER-transmembrane segment. We discuss these results in view of the conflicting interpretations in the literature. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20511.map.gz emd_20511.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20511-v30.xml emd-20511-v30.xml emd-20511.xml emd-20511.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20511_fsc.xml emd_20511_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_20511.png emd_20511.png | 57.3 KB | ||
| Filedesc metadata |  emd-20511.cif.gz emd-20511.cif.gz | 7.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20511 http://ftp.pdbj.org/pub/emdb/structures/EMD-20511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20511 | HTTPS FTP |
-Related structure data
| Related structure data |  6pwuMC  6ulcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20511.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20511.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.64 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV-1 GP160 trimer:PG16 Fab complex
| Entire | Name: HIV-1 GP160 trimer:PG16 Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 GP160 trimer:PG16 Fab complex
| Supramolecule | Name: HIV-1 GP160 trimer:PG16 Fab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / Details: recombinant protein complex |
|---|---|
| Molecular weight | Theoretical: 530 KDa |
-Supramolecule #2: HIV-1 envelope glycoprotein GP160 (mature)
| Supramolecule | Name: HIV-1 envelope glycoprotein GP160 (mature) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: mature envelope glycoprotein GP160 (i.e. cleaved form consisting of receptor binding domain GP120 and fusogenic domain GP41) |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: Antibody PG16 Fab
| Supramolecule | Name: Antibody PG16 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 97.560695 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRVRGIQRNC QHLWRWGTLI LGMLMICSAA ENLWVTVYYG VPVWKDAETT LFCASDAKAY DTEVHNVWAT HACVPTDPNP QEIYMENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPCV QLTPLCVTLD CSYNITNNIT NSITNSSVNM REEIKNCSFN M TTELRDKN ...String: MRVRGIQRNC QHLWRWGTLI LGMLMICSAA ENLWVTVYYG VPVWKDAETT LFCASDAKAY DTEVHNVWAT HACVPTDPNP QEIYMENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPCV QLTPLCVTLD CSYNITNNIT NSITNSSVNM REEIKNCSFN M TTELRDKN RKVYSLFYKL DVVQINNGNN SSNLYRLINC NTSALTQACP KVTFEPIPIR YCAPAGYAIL KCNDKEFNGT GL CKNVSTV QCTHGIRPVV STQLLLNGSL AEGKVMIRSE NITNNVKNII VQLNETVTIN CTRPNNNTRK SVRIGPGQTF YAT GDIIGD IRQAHCNVSG SQWNRALHQV VGQLREYWNT TIIFKNSSGG DLEITTHSFN CGGEFFYCNT SGLFNSNWTH NDTA SMKPN DTITLPCRIK QIINMWQRVG QAIYAPPIQG VIRCESNITG LILTRDGGGN INESQIFRPG GGDMRDNWRS ELYKY KVVR IEPLGVAPTK AKRRVVEREK RAVVELGAVF IGFLGTAGST MGAASITLTV QVRKLLSGIV QQQSNLLRAI EAQQHL LKL TVWGIKQLQA RVLAVERYLR DQQLLGIWGC SGKLICTTNV PWNSSWSNKS EREIWENMTW LQWDKEISNY THIIYEL IE ESQKQQEKNE QELLELDKWA NLWNWFDISN WLWYIKIFIM IVGGLIGLRI VFAVLSVINR VRQGYSPLSF QTLTPNPR D PDRPGRIEGE GGEQDRGRSI RLVSGFLALA WDDLRNLCLS SYHQLRDFIL IVARTVELLG HSSLKGLRLG WEGLKYLGN LLLYWGRELK TSAINLFDTI AIVVAGWTDR VIEVGQRLGR AILNIPRRIR QGLERALL UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Antibody PG16 Fab heavy chain
| Macromolecule | Name: Antibody PG16 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.952703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEFGLSWVFL VALLRGVQCQ EQLVESGGGV VQPGGSLRLS CLASGFTFHK YGMHWVRQAP GKGLEWVALI SDDGMRKYHS DSMWGRVTI SRDNSKNTLY LQFSSLKVED TAMFFCAREA GGPIWHDDVK YYDFNDGYYN YHYMDVWGKG TTVTVSSAST K GPSVFPLA ...String: MEFGLSWVFL VALLRGVQCQ EQLVESGGGV VQPGGSLRLS CLASGFTFHK YGMHWVRQAP GKGLEWVALI SDDGMRKYHS DSMWGRVTI SRDNSKNTLY LQFSSLKVED TAMFFCAREA GGPIWHDDVK YYDFNDGYYN YHYMDVWGKG TTVTVSSAST K GPSVFPLA PSSKSTSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CN VNHKPSN TKVDKRVEPK SCDKASGGGS AWSHPQFEKG GGSGGGSGGS SAWSHPQFEK |
-Macromolecule #3: Antibody PG16 Fab light chain
| Macromolecule | Name: Antibody PG16 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.424229 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAWALLLLTL LTQGTGSWAQ SALTQPASVS GSPGQTITIS CNGTSSDVGG FDSVSWYQQS PGKAPKVMVF DVSHRPSGIS NRFSGSKSG NTASLTISGL HIEDEGDYFC SSLTDRSHRI FGGGTKVTVL GQPKAAPSVT LFPPSSEELQ ANKATLVCLI S DFYPGAVT ...String: MAWALLLLTL LTQGTGSWAQ SALTQPASVS GSPGQTITIS CNGTSSDVGG FDSVSWYQQS PGKAPKVMVF DVSHRPSGIS NRFSGSKSG NTASLTISGL HIEDEGDYFC SSLTDRSHRI FGGGTKVTVL GQPKAAPSVT LFPPSSEELQ ANKATLVCLI S DFYPGAVT VAWKADSSPV KAGVETTTPS KQSNNKYAAS SYLSLTPEQW KSHKSYSCQV THEGSTVEKT VAPTECSGGS GG HHHHHHH HHH |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 44 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa Details: Quantifoil-Cu R1.2/1.3 200 mesh was glow discharged by Pelilcan EasiGlow for 30 seconds at 20 mA. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK I / Details: 4.2 second blot before plunging. | ||||||||||||
| Details | This sample was monodisperse (stoichiometry 3:1). |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 2 / Number real images: 8138 / Average exposure time: 0.125 sec. / Average electron dose: 40.0 e/Å2 Details: image size and sampling interval reported here are for super resolution. The physical pixel size of the camera is 5.0 um and image size is 3838 pixels by 3710 pixels. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus min: 0.8 µm / Calibrated magnification: 30488 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 300 |
|---|---|
| Output model |  PDB-6pwu: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)