[English] 日本語
 Yorodumi
Yorodumi- EMDB-7809: Human nuclear exosome-MTR4 RNA complex - composite map after focu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7809 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human nuclear exosome-MTR4 RNA complex - composite map after focused reconstruction | |||||||||
 Map data Map data | Composite map after focused reconstructions for a human nuclear exosome-RNA helicase complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA exosome / RNA degradation / ribonuclease / helicase / SF2 / RNA-protein complex / translocase / nuclear / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA deamination / nucleolar exosome (RNase complex) / snRNA catabolic process / positive regulation of mRNA cis splicing, via spliceosome / TRAMP complex / nuclear mRNA surveillance of mRNA 3'-end processing / nuclear polyadenylation-dependent antisense transcript catabolic process / nuclear polyadenylation-dependent snoRNA catabolic process / nuclear polyadenylation-dependent snRNA catabolic process / RNA exonuclease activity ...DNA deamination / nucleolar exosome (RNase complex) / snRNA catabolic process / positive regulation of mRNA cis splicing, via spliceosome / TRAMP complex / nuclear mRNA surveillance of mRNA 3'-end processing / nuclear polyadenylation-dependent antisense transcript catabolic process / nuclear polyadenylation-dependent snoRNA catabolic process / nuclear polyadenylation-dependent snRNA catabolic process / RNA exonuclease activity / regulation of telomerase RNA localization to Cajal body / CUT catabolic process / U1 snRNA 3'-end processing / nuclear polyadenylation-dependent CUT catabolic process / U5 snRNA 3'-end processing / TRAMP-dependent tRNA surveillance pathway / exosome (RNase complex) / mRNA decay by 3' to 5' exoribonuclease / cytoplasmic exosome (RNase complex) / U4 snRNA 3'-end processing / nuclear polyadenylation-dependent rRNA catabolic process / poly(A)-dependent snoRNA 3'-end processing / nuclear exosome (RNase complex) / exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / Tristetraprolin (TTP, ZFP36) binds and destabilizes mRNA / ATF4 activates genes in response to endoplasmic reticulum stress / Butyrate Response Factor 1 (BRF1) binds and destabilizes mRNA / positive regulation of isotype switching / histone mRNA catabolic process / nuclear mRNA surveillance / rRNA catabolic process / 7S RNA binding / mRNA 3'-UTR AU-rich region binding / isotype switching / Hydrolases; Acting on ester bonds; Endoribonucleases producing 5'-phosphomonoesters / telomerase RNA binding / RNA catabolic process / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / KSRP (KHSRP) binds and destabilizes mRNA / maturation of 5.8S rRNA / nuclear chromosome / mRNA catabolic process / negative regulation of telomere maintenance via telomerase / nuclear-transcribed mRNA catabolic process / RNA processing / Major pathway of rRNA processing in the nucleolus and cytosol / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / guanyl-nucleotide exchange factor activity / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / small-subunit processome / euchromatin / mRNA splicing, via spliceosome / fibrillar center / rRNA processing / chromosome / ribosomal small subunit biogenesis / positive regulation of cell growth / endonuclease activity / Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters / 3'-5'-RNA exonuclease activity / defense response to virus / RNA polymerase II-specific DNA-binding transcription factor binding / RNA helicase activity / single-stranded RNA binding / nuclear speck / immune response / RNA helicase / intracellular membrane-bounded organelle / nucleotide binding / DNA repair / DNA damage response / nucleolus / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / DNA binding / RNA binding / extracellular exosome / nucleoplasm / ATP binding / metal ion binding / identical protein binding / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Weick E-M / Lima CD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Helicase-Dependent RNA Decay Illuminated by a Cryo-EM Structure of a Human Nuclear RNA Exosome-MTR4 Complex. Authors: Eva-Maria Weick / M Rhyan Puno / Kurt Januszyk / John C Zinder / Michael A DiMattia / Christopher D Lima /  Abstract: The ribonucleolytic RNA exosome interacts with RNA helicases to degrade RNA. To understand how the 3' to 5' Mtr4 helicase engages RNA and the nuclear exosome, we reconstituted 14-subunit Mtr4- ...The ribonucleolytic RNA exosome interacts with RNA helicases to degrade RNA. To understand how the 3' to 5' Mtr4 helicase engages RNA and the nuclear exosome, we reconstituted 14-subunit Mtr4-containing RNA exosomes from Saccharomyces cerevisiae, Schizosaccharomyces pombe, and human and show that they unwind structured substrates to promote degradation. We loaded a human exosome with an optimized DNA-RNA chimera that stalls MTR4 during unwinding and determined its structure to an overall resolution of 3.45 Å by cryoelectron microscopy (cryo-EM). The structure reveals an RNA-engaged helicase atop the non-catalytic core, with RNA captured within the central channel and DIS3 exoribonuclease active site. MPP6 tethers MTR4 to the exosome through contacts to the RecA domains of MTR4. EXOSC10 remains bound to the core, but its catalytic module and cofactor C1D are displaced by RNA-engaged MTR4. Competition for the exosome core may ensure that RNA is committed to degradation by DIS3 when engaged by MTR4. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7809.map.gz emd_7809.map.gz | 5.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7809-v30.xml emd-7809-v30.xml emd-7809.xml emd-7809.xml | 36.4 KB 36.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7809.png emd_7809.png | 52.6 KB | ||
| Filedesc metadata |  emd-7809.cif.gz emd-7809.cif.gz | 11.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7809 http://ftp.pdbj.org/pub/emdb/structures/EMD-7809 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7809 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7809 | HTTPS FTP |
-Related structure data
| Related structure data |  6d6rMC  7808C  7810C  7812C  7813C  7814C  7815C  7818C  7819C  6d6qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7809.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7809.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map after focused reconstructions for a human nuclear exosome-RNA helicase complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
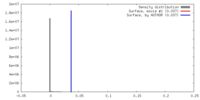
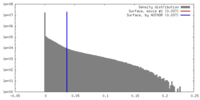
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Human nuclear exosome-MTR4 helicase captured after unwinding a DN...
+Supramolecule #1: Human nuclear exosome-MTR4 helicase captured after unwinding a DN...
+Macromolecule #1: Exosome complex component RRP45
+Macromolecule #2: Exosome complex component RRP41
+Macromolecule #3: Exosome complex component RRP43
+Macromolecule #4: Exosome complex component RRP46
+Macromolecule #5: Exosome complex component RRP42
+Macromolecule #6: Exosome complex component MTR3
+Macromolecule #7: Exosome complex component RRP40
+Macromolecule #8: Exosome complex component RRP4
+Macromolecule #9: Exosome complex component CSL4
+Macromolecule #10: Exosome component 10
+Macromolecule #11: Exosome complex exonuclease RRP44
+Macromolecule #12: M-phase phosphoprotein 6
+Macromolecule #13: Exosome RNA helicase MTR4
+Macromolecule #14: RNA (5'-R(*AP*GP*CP*AP*CP*CP*GP*UP*AP*AP*AP*GP*AP*CP*GP*C)-3')
+Macromolecule #15: DNA/RNA (62-MER)
+Macromolecule #16: ZINC ION
+Macromolecule #17: MAGNESIUM ION
+Macromolecule #18: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.037 kPa | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: 30 sec wait time, 2.5 sec blot time. | |||||||||||||||||||||
| Details | Sample was monodisperse upon elution from gel filtration prior to vitrification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 1439 / Average electron dose: 85.23 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Models were rebuilt manually using Coot, with real space refinement with local scaling followed by Phenix real space refinement with suboptimal global scaling. |
|---|---|
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Overall B value: 73.6 / Target criteria: Correlation coefficient |
| Output model |  PDB-6d6r: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)