+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20226 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Spastin hexamer in complex with substrate | |||||||||
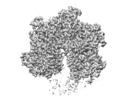
 Map data Map data | Cryo-EM map of spastin hexamer bound to substrate peptide. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ ATPase / Homohexamer / Microtubule Severing Enzyme / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule-severing ATPase / microtubule severing ATPase activity / mitotic chromosome movement towards spindle pole / microtubule severing / positive regulation of microtubule depolymerization / mitotic spindle elongation / protein hexamerization / regulation of synapse structure or activity / isomerase activity / lipid droplet ...microtubule-severing ATPase / microtubule severing ATPase activity / mitotic chromosome movement towards spindle pole / microtubule severing / positive regulation of microtubule depolymerization / mitotic spindle elongation / protein hexamerization / regulation of synapse structure or activity / isomerase activity / lipid droplet / adult locomotory behavior / spindle / nervous system development / chromosome / microtubule binding / microtubule / cell differentiation / cell division / centrosome / ATP hydrolysis activity / ATP binding / identical protein binding / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
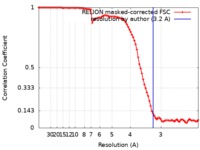
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Sandate CR / Szyk A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: An allosteric network in spastin couples multiple activities required for microtubule severing. Authors: Colby R Sandate / Agnieszka Szyk / Elena A Zehr / Gabriel C Lander / Antonina Roll-Mecak /  Abstract: The AAA+ ATPase spastin remodels microtubule arrays through severing and its mutation is the most common cause of hereditary spastic paraplegias (HSP). Polyglutamylation of the tubulin C-terminal ...The AAA+ ATPase spastin remodels microtubule arrays through severing and its mutation is the most common cause of hereditary spastic paraplegias (HSP). Polyglutamylation of the tubulin C-terminal tail recruits spastin to microtubules and modulates severing activity. Here, we present a ~3.2 Å resolution cryo-EM structure of the Drosophila melanogaster spastin hexamer with a polyglutamate peptide bound in its central pore. Two electropositive loops arranged in a double-helical staircase coordinate the substrate sidechains. The structure reveals how concurrent nucleotide and substrate binding organizes the conserved spastin pore loops into an ordered network that is allosterically coupled to oligomerization, and suggests how tubulin tail engagement activates spastin for microtubule disassembly. This allosteric coupling may apply generally in organizing AAA+ protein translocases into their active conformations. We show that this allosteric network is essential for severing and is a hotspot for HSP mutations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20226.map.gz emd_20226.map.gz | 58.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20226-v30.xml emd-20226-v30.xml emd-20226.xml emd-20226.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20226_fsc.xml emd_20226_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_20226.png emd_20226.png | 75.6 KB | ||
| Filedesc metadata |  emd-20226.cif.gz emd-20226.cif.gz | 6.7 KB | ||
| Others |  emd_20226_additional.map.gz emd_20226_additional.map.gz emd_20226_half_map_1.map.gz emd_20226_half_map_1.map.gz emd_20226_half_map_2.map.gz emd_20226_half_map_2.map.gz | 34.4 MB 56.2 MB 56.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20226 http://ftp.pdbj.org/pub/emdb/structures/EMD-20226 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20226 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20226 | HTTPS FTP |
-Validation report
| Summary document |  emd_20226_validation.pdf.gz emd_20226_validation.pdf.gz | 773.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20226_full_validation.pdf.gz emd_20226_full_validation.pdf.gz | 772.8 KB | Display | |
| Data in XML |  emd_20226_validation.xml.gz emd_20226_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_20226_validation.cif.gz emd_20226_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20226 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20226 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20226 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20226 | HTTPS FTP |
-Related structure data
| Related structure data |  6p07MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20226.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20226.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of spastin hexamer bound to substrate peptide. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map
| File | emd_20226_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_20226_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_20226_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : complex of spastin homohexamer bound to polyglutmate peptide
| Entire | Name: complex of spastin homohexamer bound to polyglutmate peptide |
|---|---|
| Components |
|
-Supramolecule #1: complex of spastin homohexamer bound to polyglutmate peptide
| Supramolecule | Name: complex of spastin homohexamer bound to polyglutmate peptide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 294 kDa/nm |
-Macromolecule #1: Spastin
| Macromolecule | Name: Spastin / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: microtubule-severing ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.183473 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPQGSKAANR PGGGYSPGPG DPLLAKQKHH HRRAFEYISK ALKIDEENEG HKELAIELYR KGIKELEDGI AVDCWSGRGD VWDRAQRLH DKMQTNLSMA RDRLHFLASG RKLTIGSKRP VNLAVANKSQ TLPRNLGSKT SVGAVQRQPV KTAATPPAVR R QFSSGRNT ...String: GPQGSKAANR PGGGYSPGPG DPLLAKQKHH HRRAFEYISK ALKIDEENEG HKELAIELYR KGIKELEDGI AVDCWSGRGD VWDRAQRLH DKMQTNLSMA RDRLHFLASG RKLTIGSKRP VNLAVANKSQ TLPRNLGSKT SVGAVQRQPV KTAATPPAVR R QFSSGRNT PPQRSRTPIN NNGPSGSGAS TPVVSVKGVE QKLVQLILDE IVEGGAKVEW TDIAGQDVAK QALQEMVILP SV RPELFTG LRAPAKGLLL FGPPGNGKTL LARAVATECS ATFLNISAAS LTSKYVGDGE KLVRALFAVA RHMQPSIIFI DQV DSLLSE RSSSEHEASR RLKTEFLVEF DGLPGNPDGD RIVVLAATNR PQELDEAALR RFTKRVYVSL PDEQTRELLL NRLL QKQGS PLDTEALRRL AKITDGYSGS DLTALAKDAA LEPIRELNVE QVKCLDISAM RAITEQDFHS SLKRIRRSVA PQSLN SYEK WSQDYGDITI UniProtKB: Spastin |
-Macromolecule #2: polyglutamate peptide
| Macromolecule | Name: polyglutamate peptide / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.954722 KDa |
| Sequence | String: EEEEEEEEEE EEEEE |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 6 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER Details: Grids were plasma-cleaned using a Solarus plasma cleaner (Gatan, Inc.) | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER Details: Sample was blotted approximately 4 seconds using Whatman No. 1 filter paper before plunge-freezing.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-48 / Number grids imaged: 2 / Number real images: 2534 / Average exposure time: 12.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)