[English] 日本語
 Yorodumi
Yorodumi- EMDB-20101: B41 SOSIP.664 trimer in complex with two Fab fragments of the sil... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20101 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | B41 SOSIP.664 trimer in complex with two Fab fragments of the silent-face targeting bNAb SF12, and one Fab fragment of the V3-targeting bNAb 10-1074 | |||||||||
 Map data Map data | Final sharpened map for class 2 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
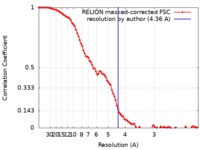
| Method | single particle reconstruction / cryo EM / Resolution: 4.36 Å | |||||||||
 Authors Authors | Barnes CO / Bjorkman PJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2019 Journal: Immunity / Year: 2019Title: Broad and Potent Neutralizing Antibodies Recognize the Silent Face of the HIV Envelope. Authors: Till Schoofs / Christopher O Barnes / Nina Suh-Toma / Jovana Golijanin / Philipp Schommers / Henning Gruell / Anthony P West / Franziska Bach / Yu Erica Lee / Lilian Nogueira / Ivelin S ...Authors: Till Schoofs / Christopher O Barnes / Nina Suh-Toma / Jovana Golijanin / Philipp Schommers / Henning Gruell / Anthony P West / Franziska Bach / Yu Erica Lee / Lilian Nogueira / Ivelin S Georgiev / Robert T Bailer / Julie Czartoski / John R Mascola / Michael S Seaman / M Juliana McElrath / Nicole A Doria-Rose / Florian Klein / Michel C Nussenzweig / Pamela J Bjorkman /   Abstract: Broadly neutralizing antibodies (bNAbs) against HIV-1 envelope (Env) inform vaccine design and are potential therapeutic agents. We identified SF12 and related bNAbs with up to 62% neutralization ...Broadly neutralizing antibodies (bNAbs) against HIV-1 envelope (Env) inform vaccine design and are potential therapeutic agents. We identified SF12 and related bNAbs with up to 62% neutralization breadth from an HIV-infected donor. SF12 recognized a glycan-dominated epitope on Env's silent face and was potent against clade AE viruses, which are poorly covered by V3-glycan bNAbs. A 3.3Å cryo-EM structure of a SF12-Env trimer complex showed additional contacts to Env protein residues by SF12 compared with VRC-PG05, the only other known donor-derived silentface antibody, explaining SF12's increased neutralization breadth, potency, and resistance to Env mutation routes. Asymmetric binding of SF12 was associated with distinct N-glycan conformations across Env protomers, demonstrating intra-Env glycan heterogeneity. Administrating SF12 to HIV-1-infected humanized mice suppressed viremia and selected for viruses lacking the N448 glycan. Effective bNAbs can therefore be raised against HIV-1 Env's silent face, suggesting their potential for HIV-1 prevention, therapy, and vaccine development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20101.map.gz emd_20101.map.gz | 10.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20101-v30.xml emd-20101-v30.xml emd-20101.xml emd-20101.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20101_fsc.xml emd_20101_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20101.png emd_20101.png | 144.8 KB | ||
| Others |  emd_20101_additional.map.gz emd_20101_additional.map.gz emd_20101_half_map_1.map.gz emd_20101_half_map_1.map.gz emd_20101_half_map_2.map.gz emd_20101_half_map_2.map.gz | 48.9 MB 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20101 http://ftp.pdbj.org/pub/emdb/structures/EMD-20101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20101 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20101.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20101.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final sharpened map for class 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened refined map
| File | emd_20101_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened refined map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_20101_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_20101_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of B41 SOSIP.664 trimer with three SF12 Fabs and one 10-1...
| Entire | Name: Complex of B41 SOSIP.664 trimer with three SF12 Fabs and one 10-1074 Fab. |
|---|---|
| Components |
|
-Supramolecule #1: Complex of B41 SOSIP.664 trimer with three SF12 Fabs and one 10-1...
| Supramolecule | Name: Complex of B41 SOSIP.664 trimer with three SF12 Fabs and one 10-1074 Fab. type: complex / ID: 1 / Parent: 0 Details: Fab fragment generated by recombinant expression and complexed with the B41 SOSIP.664 trimer |
|---|---|
| Molecular weight | Theoretical: 540 KDa |
-Supramolecule #2: B41 SOSIP.664 trimer
| Supramolecule | Name: B41 SOSIP.664 trimer / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  |
-Supramolecule #3: 10-1074 Fab
| Supramolecule | Name: 10-1074 Fab / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant strain: Expi293 / Recombinant plasmid: p3BNC Homo sapiens (human) / Recombinant strain: Expi293 / Recombinant plasmid: p3BNC |
-Supramolecule #4: SF12 Fab
| Supramolecule | Name: SF12 Fab / type: complex / ID: 4 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant strain: Expi293 / Recombinant plasmid: p3BNC Homo sapiens (human) / Recombinant strain: Expi293 / Recombinant plasmid: p3BNC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Details: unspecified | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV Details: 0 blot force, 3 second blot time, 3 uL sample added to freshly glow-discharged grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2732 / Average exposure time: 8.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)