+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20025 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Eastern equine encephalitis virus (EEEV) | |||||||||
 Map data Map data | native EEEV reconstruction | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Eastern equine encephalitis virus Eastern equine encephalitis virus | |||||||||
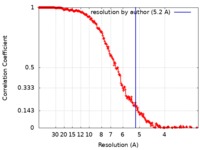
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | |||||||||
 Authors Authors | Rossmann MG / Chen CL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Cryo-EM structure of eastern equine encephalitis virus in complex with heparan sulfate analogues. Authors: Chun-Liang Chen / S Saif Hasan / Thomas Klose / Yingyuan Sun / Geeta Buda / Chengqun Sun / William B Klimstra / Michael G Rossmann /  Abstract: Eastern equine encephalitis virus (EEEV), a mosquito-borne icosahedral alphavirus found mainly in North America, causes human and equine neurotropic infections. EEEV neurovirulence is influenced by ...Eastern equine encephalitis virus (EEEV), a mosquito-borne icosahedral alphavirus found mainly in North America, causes human and equine neurotropic infections. EEEV neurovirulence is influenced by the interaction of the viral envelope protein E2 with heparan sulfate (HS) proteoglycans from the host's plasma membrane during virus entry. Here, we present a 5.8-Å cryoelectron microscopy (cryo-EM) structure of EEEV complexed with the HS analog heparin. "Peripheral" HS binding sites were found to be associated with the base of each of the E2 glycoproteins that form the 60 quasi-threefold spikes (q3) and the 20 sites associated with the icosahedral threefold axes (i3). In addition, there is one HS site at the vertex of each q3 and i3 spike (the "axial" sites). Both the axial and peripheral sites are surrounded by basic residues, suggesting an electrostatic mechanism for HS binding. These residues are highly conserved among EEEV strains, and therefore a change in these residues might be linked to EEEV neurovirulence. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20025.map.gz emd_20025.map.gz | 348.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20025-v30.xml emd-20025-v30.xml emd-20025.xml emd-20025.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20025_fsc.xml emd_20025_fsc.xml | 21.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_20025.png emd_20025.png | 159.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20025 http://ftp.pdbj.org/pub/emdb/structures/EMD-20025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20025 | HTTPS FTP |
-Validation report
| Summary document |  emd_20025_validation.pdf.gz emd_20025_validation.pdf.gz | 78.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20025_full_validation.pdf.gz emd_20025_full_validation.pdf.gz | 77.2 KB | Display | |
| Data in XML |  emd_20025_validation.xml.gz emd_20025_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20025 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20025 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20025 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20025 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20025.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20025.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | native EEEV reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.58 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EEEV
| Entire | Name: EEEV |
|---|---|
| Components |
|
-Supramolecule #1: EEEV
| Supramolecule | Name: EEEV / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Recombinant expression | Organism:  Cricetinae gen. sp. (mammal) / Recombinant cell: BHK Cricetinae gen. sp. (mammal) / Recombinant cell: BHK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa / Details: unspecified | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)