[English] 日本語
 Yorodumi
Yorodumi- EMDB-19724: RNA polymerase II early elongation complex bound to TFIIE, TFIIF ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA polymerase II early elongation complex bound to TFIIE, TFIIF and TFIIH - state a | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase II / promoter escape / de novo transcription / TRANSCRIPTION | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Zhan Y / Grabbe F / Oberbeckmann E / Dienemann C / Cramer P | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Three-step mechanism of promoter escape by RNA polymerase II. Authors: Yumeng Zhan / Frauke Grabbe / Elisa Oberbeckmann / Christian Dienemann / Patrick Cramer /  Abstract: The transition from transcription initiation to elongation is highly regulated in human cells but remains incompletely understood at the structural level. In particular, it is unclear how ...The transition from transcription initiation to elongation is highly regulated in human cells but remains incompletely understood at the structural level. In particular, it is unclear how interactions between RNA polymerase II (RNA Pol II) and initiation factors are broken to enable promoter escape. Here, we reconstitute RNA Pol II promoter escape in vitro and determine high-resolution structures of initially transcribing complexes containing 8-, 10-, and 12-nt ordered RNAs and two elongation complexes containing 14-nt RNAs. We suggest that promoter escape occurs in three major steps. First, the growing RNA displaces the B-reader element of the initiation factor TFIIB without evicting TFIIB. Second, the rewinding of the transcription bubble coincides with the eviction of TFIIA, TFIIB, and TBP. Third, the binding of DSIF and NELF facilitates TFIIE and TFIIH dissociation, establishing the paused elongation complex. This three-step model for promoter escape fills a gap in our understanding of the initiation-elongation transition of RNA Pol II transcription. | |||||||||
| History |
|
- Structure visualization
Structure visualization
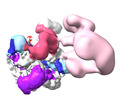
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19724.map.gz emd_19724.map.gz | 140 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19724-v30.xml emd-19724-v30.xml emd-19724.xml emd-19724.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19724.png emd_19724.png | 84.2 KB | ||
| Masks |  emd_19724_msk_1.map emd_19724_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19724.cif.gz emd-19724.cif.gz | 3.7 KB | ||
| Others |  emd_19724_half_map_1.map.gz emd_19724_half_map_1.map.gz emd_19724_half_map_2.map.gz emd_19724_half_map_2.map.gz | 193.7 MB 193.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19724 http://ftp.pdbj.org/pub/emdb/structures/EMD-19724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19724 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19724.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19724.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
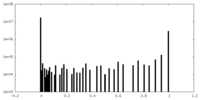
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
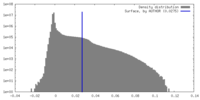
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19724_msk_1.map emd_19724_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
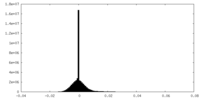
| Density Histograms |
-Half map: #1
| File | emd_19724_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
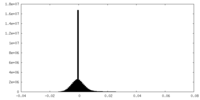
| Density Histograms |
-Half map: #2
| File | emd_19724_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mammalian RNA polymerase II general transcription factor IIE gene...
| Entire | Name: mammalian RNA polymerase II general transcription factor IIE general transcription factor IIF general transcription factor IIH DNA |
|---|---|
| Components |
|
-Supramolecule #1: mammalian RNA polymerase II general transcription factor IIE gene...
| Supramolecule | Name: mammalian RNA polymerase II general transcription factor IIE general transcription factor IIF general transcription factor IIH DNA type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: RNA polymerase II
| Supramolecule | Name: RNA polymerase II / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 12628 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)