+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1969 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of HBV Cp183 capsid | |||||||||
 Map data Map data | This is a cryo-EM 3D map of HBV Cp183 capsid | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HBV / Cp183 | |||||||||
| Biological species |   Hepatitis B virus Hepatitis B virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 17.4 Å | |||||||||
 Authors Authors | Chen C / Wang JC-Y / Zlotnick A | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2011 Journal: PLoS Pathog / Year: 2011Title: A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. Authors: Chao Chen / Joseph Che-Yen Wang / Adam Zlotnick /  Abstract: The C-terminal domain (CTD) of Hepatitis B virus (HBV) core protein is involved in regulating multiple stages of the HBV lifecycle. CTD phosphorylation correlates with pregenomic-RNA encapsidation ...The C-terminal domain (CTD) of Hepatitis B virus (HBV) core protein is involved in regulating multiple stages of the HBV lifecycle. CTD phosphorylation correlates with pregenomic-RNA encapsidation during capsid assembly, reverse transcription, and viral transport, although the mechanisms remain unknown. In vitro, purified HBV core protein (Cp183) binds any RNA and assembles aggressively, independent of phosphorylation, to form empty and RNA-filled capsids. We hypothesize that there must be a chaperone that binds the CTD to prevent self-assembly and nonspecific RNA packaging. Here, we show that HBV capsid assembly is stalled by the Serine Arginine protein kinase (SRPK) binding to the CTD, and reactivated by subsequent phosphorylation. Using the SRPK to probe capsids, solution and structural studies showed that SRPK bound to capsid, though the CTD is sequestered on the capsid interior. This result indicates transient CTD externalization and suggests that capsid dynamics could be crucial for directing HBV intracellular trafficking. Our studies illustrate the stochastic nature of virus capsids and demonstrate the appropriation of a host protein by a virus for a non-canonical function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1969.map.gz emd_1969.map.gz | 9.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1969-v30.xml emd-1969-v30.xml emd-1969.xml emd-1969.xml | 8.5 KB 8.5 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1969.png EMD-1969.png | 119.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1969 http://ftp.pdbj.org/pub/emdb/structures/EMD-1969 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1969 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1969 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1969.map.gz / Format: CCP4 / Size: 30.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1969.map.gz / Format: CCP4 / Size: 30.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a cryo-EM 3D map of HBV Cp183 capsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.94 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
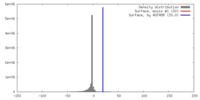
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HBV Cp183 capsid
| Entire | Name: HBV Cp183 capsid |
|---|---|
| Components |
|
-Supramolecule #1000: HBV Cp183 capsid
| Supramolecule | Name: HBV Cp183 capsid / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Hepatitis B virus
| Supramolecule | Name: Hepatitis B virus / type: virus / ID: 1 / Name.synonym: HBV Cp183 / Details: HBV Cp183 was purified from E. coli / NCBI-ID: 10407 / Sci species name: Hepatitis B virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes / Syn species name: HBV Cp183 |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Virus shell | Shell ID: 1 / Name: Cp183 / T number (triangulation number): 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.24 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 0.53 M NaCl, 10 mM DTT, 20 mM Tris-HCl |
| Grid | Details: Quantifoil R 2/2 holey carbon 200 mesh copper grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 93 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot / Method: Blot for 4 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FS |
|---|---|
| Temperature | Average: 97 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 80,000 times magnification |
| Specialist optics | Energy filter - Name: Omega filter |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 19 / Average electron dose: 14 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.1 mm / Nominal defocus max: 2.09 µm / Nominal defocus min: 1.23 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle phase-flipping |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 17.4 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Auto3dem / Number images used: 955 |
 Movie
Movie Controller
Controller




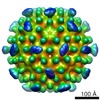










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)