+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12252 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | pT=7 laevo quasi-symmetric bacterial microcompartment particle | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Kalnins G / Cesle EE | |||||||||
| Funding support |  Latvia, Latvia,  Czech Republic, 2 items Czech Republic, 2 items
| |||||||||
 Citation Citation |  Journal: Protein Sci / Year: 2021 Journal: Protein Sci / Year: 2021Title: Variety of size and form of GRM2 bacterial microcompartment particles. Authors: Eva Emilija Cesle / Anatolij Filimonenko / Kaspars Tars / Gints Kalnins /   Abstract: Bacterial microcompartments (BMCs) are bacterial organelles involved in enzymatic processes, such as carbon fixation, choline, ethanolamine and propanediol degradation, and others. Formed of a semi- ...Bacterial microcompartments (BMCs) are bacterial organelles involved in enzymatic processes, such as carbon fixation, choline, ethanolamine and propanediol degradation, and others. Formed of a semi-permeable protein shell and an enzymatic core, they can enhance enzyme performance and protect the cell from harmful intermediates. With the ability to encapsulate non-native enzymes, BMCs show high potential for applied use. For this goal, a detailed look into shell form variability is significant to predict shell adaptability. Here we present four novel 3D cryo-EM maps of recombinant Klebsiella pneumoniae GRM2 BMC shell particles with the resolution in range of 9 to 22 Å and nine novel 2D classes corresponding to discrete BMC shell forms. These structures reveal icosahedral, elongated, oblate, multi-layered and polyhedral traits of BMCs, indicating considerable variation in size and form as well as adaptability during shell formation processes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12252.map.gz emd_12252.map.gz | 80.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12252-v30.xml emd-12252-v30.xml emd-12252.xml emd-12252.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
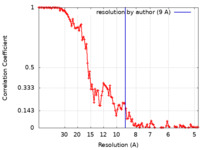
| FSC (resolution estimation) |  emd_12252_fsc.xml emd_12252_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_12252.png emd_12252.png | 82.8 KB | ||
| Others |  emd_12252_half_map_1.map.gz emd_12252_half_map_1.map.gz emd_12252_half_map_2.map.gz emd_12252_half_map_2.map.gz | 80.9 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12252 http://ftp.pdbj.org/pub/emdb/structures/EMD-12252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12252 | HTTPS FTP |
-Validation report
| Summary document |  emd_12252_validation.pdf.gz emd_12252_validation.pdf.gz | 309.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12252_full_validation.pdf.gz emd_12252_full_validation.pdf.gz | 308.9 KB | Display | |
| Data in XML |  emd_12252_validation.xml.gz emd_12252_validation.xml.gz | 16.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12252 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12252 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12252 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12252 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12252.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12252.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.44 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_12252_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
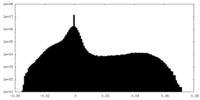
| Projections & Slices |
| ||||||||||||
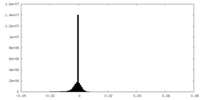
| Density Histograms |
-Half map: #1
| File | emd_12252_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
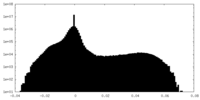
| Projections & Slices |
| ||||||||||||
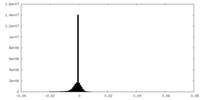
| Density Histograms |
- Sample components
Sample components
-Entire : pT=7 laevo quasi-symmetric bacterial microcompartment particles
| Entire | Name: pT=7 laevo quasi-symmetric bacterial microcompartment particles |
|---|---|
| Components |
|
-Supramolecule #1: pT=7 laevo quasi-symmetric bacterial microcompartment particles
| Supramolecule | Name: pT=7 laevo quasi-symmetric bacterial microcompartment particles type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: bacterial microcompartment particle obtained by recombinant expression of pentameric EutN and hexameric cmcC subunits |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Recombinant expression | Organism:  |
-Macromolecule #1: cmcC/PduA/ccmK BMC-H bacterial microcompartment protein
| Macromolecule | Name: cmcC/PduA/ccmK BMC-H bacterial microcompartment protein type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MKEALGLIET KGLVACIEAA DAMCKAANVE LIGYENVGSG LVTAMVKGDV GAVNAAVDSG VEAAKRIGKV VSSRVIARPH NDIEKIAGST KHKSLRPHNA |
-Macromolecule #2: cmcD/EutN BMC-P bacterial microcompartment protein
| Macromolecule | Name: cmcD/EutN BMC-P bacterial microcompartment protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MILAKVTGHV VATQKCDELR GSNLLLITRL DDKQQPMKDQ TWVAVDNVGA GMHDIVLAEE YFALNKDRYK AMSVVAIVEK VFRDTEQE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 / Component:
| ||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4s before plunging. | ||||||
| Details | Sample was purified with ultracentrifugation and gel filtration |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 3240 / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.7000000000000001 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)