[English] 日本語
 Yorodumi
Yorodumi- EMDB-19247: NDH-PSI-LHCI supercomplex from S. oleracea: local refined border ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | NDH-PSI-LHCI supercomplex from S. oleracea: local refined border region between NDH and PSI-LHCI-2 | |||||||||
 Map data Map data | filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NDHI / PSI / Supercomplex / photosynthesis / electron transport chain / lipids / proton translocation / plastoquinone / ELECTRON TRANSPORT | |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
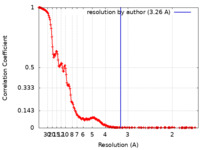
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Introini B / Hahn A / Kuehlbrandt W | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Cryo-EM structure of the NDH-PSI-LHCI supercomplex from Spinacia oleracea. Authors: Bianca Introini / Alexander Hahn / Werner Kühlbrandt /  Abstract: The nicotinamide adenine dinucleotide phosphate (NADPH) dehydrogenase (NDH) complex is crucial for photosynthetic cyclic electron flow and respiration, transferring electrons from ferredoxin to ...The nicotinamide adenine dinucleotide phosphate (NADPH) dehydrogenase (NDH) complex is crucial for photosynthetic cyclic electron flow and respiration, transferring electrons from ferredoxin to plastoquinone while transporting H across the chloroplast membrane. This process boosts adenosine triphosphate production, regardless of NADPH levels. In flowering plants, NDH forms a supercomplex with photosystem I, enhancing its stability under high light. We report the cryo-electron microscopy structure of the NDH supercomplex in Spinacia oleracea at a resolution of 3.0-3.3 Å. The supercomplex consists of 41 protein subunits, 154 chlorophylls and 38 carotenoids. Subunit interactions are reinforced by 46 distinct lipids. The structure of NDH resembles that of mitochondrial complex I closely, including the quinol-binding site and an extensive internal aqueous passage for proton translocation. A well-resolved catalytic plastoquinone (PQ) occupies the PQ channel. The pronounced structural similarity to complex I sheds light on electron transfer and proton translocation within the NDH supercomplex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19247.map.gz emd_19247.map.gz | 483.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19247-v30.xml emd-19247-v30.xml emd-19247.xml emd-19247.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19247_fsc.xml emd_19247_fsc.xml | 23.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_19247.png emd_19247.png | 77.3 KB | ||
| Masks |  emd_19247_msk_1.map emd_19247_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19247.cif.gz emd-19247.cif.gz | 4.3 KB | ||
| Others |  emd_19247_additional_1.map.gz emd_19247_additional_1.map.gz emd_19247_half_map_1.map.gz emd_19247_half_map_1.map.gz emd_19247_half_map_2.map.gz emd_19247_half_map_2.map.gz | 255.2 MB 474.9 MB 474.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19247 http://ftp.pdbj.org/pub/emdb/structures/EMD-19247 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19247 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19247 | HTTPS FTP |
-Validation report
| Summary document |  emd_19247_validation.pdf.gz emd_19247_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19247_full_validation.pdf.gz emd_19247_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_19247_validation.xml.gz emd_19247_validation.xml.gz | 25.4 KB | Display | |
| Data in CIF |  emd_19247_validation.cif.gz emd_19247_validation.cif.gz | 34.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19247 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19247 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19247 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19247 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19247.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19247.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19247_msk_1.map emd_19247_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unfiltered map
| File | emd_19247_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_19247_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_19247_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NDHI-PSI supercomplex
| Entire | Name: NDHI-PSI supercomplex |
|---|---|
| Components |
|
-Supramolecule #1: NDHI-PSI supercomplex
| Supramolecule | Name: NDHI-PSI supercomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#42 |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)