+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-1892 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
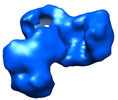
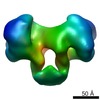
| タイトル | EcoR124 Type I DNA restriction-modification enzyme complex (in closed state) with bound DNA mimic protein Ocr from phage T7. 3D reconstruction by single particle analysis from negative stain EM. | |||||||||
 マップデータ マップデータ | 3D reconstruction of EcoR124I Type I restriction enzyme complex with a bound antirestriction protein Ocr. | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | EcoR124 / endonuclease / methyltransferase / type I restriction / DNA mimic / HsdS / HsdM / HsdR / electron microscopy / negative stain / translocase / DEAD-box / ATPase / antirestriction / phage / T7 | |||||||||
| 機能・相同性 | Protein Ocr / Restriction endonuclease, type I, HsdR / Type I restriction modification DNA specificity domain / DNA methylase, adenine-specific / protein binding / DNA restriction-modification system 機能・相同性情報 機能・相同性情報 | |||||||||
| 生物種 |    Enterobacteria phage T7 (ファージ) Enterobacteria phage T7 (ファージ) | |||||||||
| 手法 | 単粒子再構成法 / ネガティブ染色法 / 解像度: 24.0 Å | |||||||||
 データ登録者 データ登録者 | Kennaway CK / Taylor JE / Song CF / Potrzebowski W / White JH / Swiderska A / Obarska-Kosinska A / Callow P / Cooper LP / Roberts GA ...Kennaway CK / Taylor JE / Song CF / Potrzebowski W / White JH / Swiderska A / Obarska-Kosinska A / Callow P / Cooper LP / Roberts GA / Bujnicki JM / Trinick J / Kneale GG / Dryden DTF | |||||||||
 引用 引用 |  ジャーナル: Genes Dev / 年: 2012 ジャーナル: Genes Dev / 年: 2012タイトル: Structure and operation of the DNA-translocating type I DNA restriction enzymes. 著者: Christopher K Kennaway / James E N Taylor / Chun Feng Song / Wojciech Potrzebowski / William Nicholson / John H White / Anna Swiderska / Agnieszka Obarska-Kosinska / Philip Callow / Laurie P ...著者: Christopher K Kennaway / James E N Taylor / Chun Feng Song / Wojciech Potrzebowski / William Nicholson / John H White / Anna Swiderska / Agnieszka Obarska-Kosinska / Philip Callow / Laurie P Cooper / Gareth A Roberts / Jean-Baptiste Artero / Janusz M Bujnicki / John Trinick / G Geoff Kneale / David T F Dryden /    要旨: Type I DNA restriction/modification (RM) enzymes are molecular machines found in the majority of bacterial species. Their early discovery paved the way for the development of genetic engineering. ...Type I DNA restriction/modification (RM) enzymes are molecular machines found in the majority of bacterial species. Their early discovery paved the way for the development of genetic engineering. They control (restrict) the influx of foreign DNA via horizontal gene transfer into the bacterium while maintaining sequence-specific methylation (modification) of host DNA. The endonuclease reaction of these enzymes on unmethylated DNA is preceded by bidirectional translocation of thousands of base pairs of DNA toward the enzyme. We present the structures of two type I RM enzymes, EcoKI and EcoR124I, derived using electron microscopy (EM), small-angle scattering (neutron and X-ray), and detailed molecular modeling. DNA binding triggers a large contraction of the open form of the enzyme to a compact form. The path followed by DNA through the complexes is revealed by using a DNA mimic anti-restriction protein. The structures reveal an evolutionary link between type I RM enzymes and type II RM enzymes. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_1892.map.gz emd_1892.map.gz | 253.7 KB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-1892-v30.xml emd-1892-v30.xml emd-1892.xml emd-1892.xml | 15.1 KB 15.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_1892.png emd_1892.png | 217.6 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1892 http://ftp.pdbj.org/pub/emdb/structures/EMD-1892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1892 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_1892_validation.pdf.gz emd_1892_validation.pdf.gz | 194.7 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_1892_full_validation.pdf.gz emd_1892_full_validation.pdf.gz | 193.8 KB | 表示 | |
| XML形式データ |  emd_1892_validation.xml.gz emd_1892_validation.xml.gz | 5.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1892 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1892 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_1892.map.gz / 形式: CCP4 / 大きさ: 1.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_1892.map.gz / 形式: CCP4 / 大きさ: 1.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | 3D reconstruction of EcoR124I Type I restriction enzyme complex with a bound antirestriction protein Ocr. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 3.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : EcoR124I R2 M2 S1 complex with dimeric Ocr bound at target recogn...
| 全体 | 名称: EcoR124I R2 M2 S1 complex with dimeric Ocr bound at target recognition domains |
|---|---|
| 要素 |
|
-超分子 #1000: EcoR124I R2 M2 S1 complex with dimeric Ocr bound at target recogn...
| 超分子 | 名称: EcoR124I R2 M2 S1 complex with dimeric Ocr bound at target recognition domains タイプ: sample / ID: 1000 / 詳細: Stained with uranyl acetate / 集合状態: 1x HsdS, 2x HsdM, 2x HsdR, 2x Ocr / Number unique components: 4 |
|---|---|
| 分子量 | 理論値: 428 KDa |
-分子 #1: EcoR124I HsdS specificity subunit
| 分子 | 名称: EcoR124I HsdS specificity subunit / タイプ: protein_or_peptide / ID: 1 / Name.synonym: HsdS / コピー数: 1 / 集合状態: Monomer / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 50 KDa |
| 配列 | GO: protein binding InterPro: Type I restriction modification DNA specificity domain |
-分子 #2: EcoR124I HsdM methyltransferase subunit
| 分子 | 名称: EcoR124I HsdM methyltransferase subunit / タイプ: protein_or_peptide / ID: 2 / Name.synonym: HsdM / コピー数: 2 / 集合状態: Dimer / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 59 KDa |
| 配列 | GO: protein binding / InterPro: DNA methylase, adenine-specific |
-分子 #3: EcoR124I HsdR endonuclease subunit
| 分子 | 名称: EcoR124I HsdR endonuclease subunit / タイプ: protein_or_peptide / ID: 3 / Name.synonym: HsdR / コピー数: 2 / 集合状態: Monomer / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 120 KDa |
| 配列 | GO: DNA restriction-modification system / InterPro: Restriction endonuclease, type I, HsdR |
-分子 #4: ORF 0.3 Ocr antirestriction protein
| 分子 | 名称: ORF 0.3 Ocr antirestriction protein / タイプ: protein_or_peptide / ID: 4 / Name.synonym: Ocr 詳細: Ocr forms a dimer that mimics B-form DNA in shape and charge distrubution. コピー数: 2 / 集合状態: Dimer / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:   Enterobacteria phage T7 (ファージ) / 別称: T7 Enterobacteria phage T7 (ファージ) / 別称: T7 |
| 分子量 | 理論値: 27 KDa |
| 組換発現 | 生物種:  |
| 配列 | InterPro: Protein Ocr |
-実験情報
-構造解析
| 手法 | ネガティブ染色法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.050 mg/mL |
|---|---|
| 緩衝液 | pH: 4.7 / 詳細: 20mM Tris-Cl, 100 mM NaCl |
| 染色 | タイプ: NEGATIVE 詳細: Protein was adsorbed onto UV treated carbon for 1 minute, blotted, then 1% uranyl acetate solution was applied for 1 min then blotted, three times. |
| グリッド | 詳細: 400 mesh copper, continuous carbon |
| 凍結 | 凍結剤: NONE / 装置: OTHER / 詳細: Negative stain |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | JEOL 1200EXII |
|---|---|
| 温度 | 平均: 294 K |
| アライメント法 | Legacy - 非点収差: Corrected at 80,000x |
| 詳細 | Customised JEOL 1200 EX microscope, low dose mode. |
| 日付 | 2011年1月13日 |
| 撮影 | カテゴリ: FILM / フィルム・検出器のモデル: KODAK SO-163 FILM / デジタル化 - スキャナー: OTHER / デジタル化 - サンプリング間隔: 15 µm / 実像数: 7 / 平均電子線量: 40 e/Å2 / 詳細: Scanned on Imacon scanner / ビット/ピクセル: 8 |
| 電子線 | 加速電圧: 80 kV / 電子線源: TUNGSTEN HAIRPIN |
| 電子光学系 | 倍率(補正後): 37833 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.0 mm / 最大 デフォーカス(公称値): 1.357 µm / 最小 デフォーカス(公称値): 0.391 µm / 倍率(公称値): 40000 |
| 試料ステージ | 試料ホルダー: Side entry / 試料ホルダーモデル: JEOL |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | PDB ID: Chain - Chain ID: A |
|---|---|
| ソフトウェア | 名称:  Chimera Chimera |
| 詳細 | PDBEntryID_givenInChain. Protocol: rigid body. HsdR (2W00) was fitted into the density after the core methylase (HsdS and 2x HsdM) was fitted. Ocr docked into DNA binding sites (target recognition domains) in the methylase. |
| 精密化 | 空間: RECIPROCAL / プロトコル: RIGID BODY FIT / 当てはまり具合の基準: cross-correlation |
 ムービー
ムービー コントローラー
コントローラー










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















