[English] 日本語
 Yorodumi
Yorodumi- EMDB-18559: C1 turret to capsid interface of full Haloferax tailed virus 1 ad... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C1 turret to capsid interface of full Haloferax tailed virus 1 adjacent to the portal-capsid interface. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Archaeal virus / turret / turret capsid interface / Mg ions / VIRUS | |||||||||
| Function / homology | NodB homology domain / Polysaccharide deacetylase / hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds / Glycoside hydrolase/deacetylase, beta/alpha-barrel / carbohydrate metabolic process / HK97 gp5-like major capsid protein / Prokaryotic polysaccharide deacetylase Function and homology information Function and homology information | |||||||||
| Biological species |  Haloferax tailed virus 1 Haloferax tailed virus 1 | |||||||||
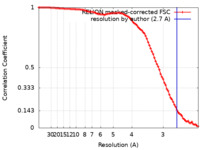
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Zhang D / Daum B / Isupov MN / McLaren M | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2025 Journal: Sci Adv / Year: 2025Title: Cryo-EM resolves the structure of the archaeal dsDNA virus HFTV1 from head to tail. Authors: Daniel X Zhang / Michail N Isupov / Rebecca M Davies / Sabine Schwarzer / Mathew McLaren / William S Stuart / Vicki A M Gold / Hanna M Oksanen / Tessa E F Quax / Bertram Daum /    Abstract: While archaeal viruses show a stunning diversity of morphologies, many bear a notable resemblance to tailed bacterial phages. This raises fundamental questions: Do all tailed viruses share a common ...While archaeal viruses show a stunning diversity of morphologies, many bear a notable resemblance to tailed bacterial phages. This raises fundamental questions: Do all tailed viruses share a common origin and do they infect their hosts in similar ways? Answering these questions requires high-resolution structural insights, yet no complete atomic models of archaeal viruses have been available. Here, we present the near-atomic resolution structure of Haloferax tailed virus 1 (HFTV1), an archaeal virus thriving in extreme salinity. Using cryo-electron microscopy, we resolve the architecture and assembly of all structural proteins and capture conformational transitions associated with DNA ejection. Our data reveal genome spooling within the capsid and identify putative receptor-binding and catalytic sites for host recognition and infection. These findings uncover key mechanisms of archaeal virus assembly, principles of virus-host interactions, and evolutionary links connecting archaeal, bacterial, and eukaryotic viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18559.map.gz emd_18559.map.gz | 48.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18559-v30.xml emd-18559-v30.xml emd-18559.xml emd-18559.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18559_fsc.xml emd_18559_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_18559.png emd_18559.png | 82.4 KB | ||
| Filedesc metadata |  emd-18559.cif.gz emd-18559.cif.gz | 6.6 KB | ||
| Others |  emd_18559_additional_1.map.gz emd_18559_additional_1.map.gz emd_18559_half_map_1.map.gz emd_18559_half_map_1.map.gz emd_18559_half_map_2.map.gz emd_18559_half_map_2.map.gz | 88.4 MB 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18559 http://ftp.pdbj.org/pub/emdb/structures/EMD-18559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18559 | HTTPS FTP |
-Related structure data
| Related structure data |  8qpqMC  8qpgC  8qqnC  8qsiC  8qsyC  9fkbC  9gs0C  9h4pC  9h5bC  9h7vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18559.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18559.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.171 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_18559_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18559_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18559_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Haloferax tailed virus 1
| Entire | Name:  Haloferax tailed virus 1 Haloferax tailed virus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Haloferax tailed virus 1
| Supramolecule | Name: Haloferax tailed virus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / NCBI-ID: 2507575 / Sci species name: Haloferax tailed virus 1 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Haloferax gibbonsii (archaea) Haloferax gibbonsii (archaea) |
-Macromolecule #1: Prokaryotic polysaccharide deacetylase
| Macromolecule | Name: Prokaryotic polysaccharide deacetylase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloferax tailed virus 1 Haloferax tailed virus 1 |
| Molecular weight | Theoretical: 45.242082 KDa |
| Sequence | String: MTGLNPDGLG RTAAFSNTSA ESVSAVDATI DRLYAQDRIE IPTDSRQLFS TRGTVLRNFE DLSGWTANIG SLSAETSDVY VGSQSARLT ASSSAVDIRY SFGTAQDFTG KGFSMALKRI DVSGSSDSTP IKIRLVDGNT NYRTFSARCR PGGGDEWGRR D FGFESEDT ...String: MTGLNPDGLG RTAAFSNTSA ESVSAVDATI DRLYAQDRIE IPTDSRQLFS TRGTVLRNFE DLSGWTANIG SLSAETSDVY VGSQSARLT ASSSAVDIRY SFGTAQDFTG KGFSMALKRI DVSGSSDSTP IKIRLVDGNT NYRTFSARCR PGGGDEWGRR D FGFESEDT GFDVTNVQTM TVTTNSRSSI DILVDDIRVV DSSGTGQVIV TIDDVHTGDK TAAEVFGRYG IPIGLAANAK FL DQSSSKL TTQEFKDLLA KPHVYAVNHG YNHYDYGSYS IDEIEDDVIR GKYELQDLGV REPNINHYVY PSGNYAQESI DML SNYHVM SWGTGAESFD ALTPNQLTSP WHNLRCSFDS GTAEAEQAVN DAATYNQTAH IYFHSDNVTQ SEMESVAQTI NSAD VTPIT LMDFYNQQ UniProtKB: Prokaryotic polysaccharide deacetylase |
-Macromolecule #2: gp30
| Macromolecule | Name: gp30 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloferax tailed virus 1 Haloferax tailed virus 1 |
| Molecular weight | Theoretical: 12.005731 KDa |
| Sequence | String: MTDTIVNVQG SFFSASASGV ADTESLLIDP QDAKFGAIEI HNIA(NEP)GGSVD VELLTSSDDT ELVEDAAVTL DSFTGE GIS QGNQIEASDN TNTYIRITNT SGGAIDIIAT GREVSQ |
-Macromolecule #3: HK97 gp5-like major capsid protein
| Macromolecule | Name: HK97 gp5-like major capsid protein / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloferax tailed virus 1 Haloferax tailed virus 1 |
| Molecular weight | Theoretical: 43.544406 KDa |
| Sequence | String: MLMEAALPGS DVSAREVAKV WPGAKKGDYS FLQGNQSRSL EAEMTRTARA EAGTDRHRAL KDYAVDADNL PKTLSAGSKH LTEDGDVIE ARLDDAIPRM LFAASDPEYV DTLFREQLLE VVMEGRELRK VAREASNVIN ANTRVGDVPI ASDEEFARPT G QGAEIRDD ...String: MLMEAALPGS DVSAREVAKV WPGAKKGDYS FLQGNQSRSL EAEMTRTARA EAGTDRHRAL KDYAVDADNL PKTLSAGSKH LTEDGDVIE ARLDDAIPRM LFAASDPEYV DTLFREQLLE VVMEGRELRK VAREASNVIN ANTRVGDVPI ASDEEFARPT G QGAEIRDD GETYTTVAWN ATKLTEGSRV TDEMRDQAMV DLIERNIQRV GASLENGINR VFLTELVDNA QNNHDTAGSN QG YQALNSA VGEVDKDDFR PDTYVTHPDY RTQLFNDTNL AYANRAGTNE VLRNREDAPI VGDIAGLDMH AAMSSATYDD GTD IGWSGG SETWGFSSDG DKGAVVYDRD NIHTILYAPN GQDVEIKDYE DPIRDITGVN GRLHVDCQYS QGRSSATVQY UniProtKB: HK97 gp5-like major capsid protein |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 50 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 54.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8qpq: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)