[English] 日本語
 Yorodumi
Yorodumi- EMDB-1840: The RNA polymerase and transcription elongation factor Spt4-5 com... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1840 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The RNA polymerase and transcription elongation factor Spt4-5 complex structure | |||||||||
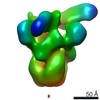
 Map data Map data | ccp4 density map for PFu RNA polymerase-Spt4-Spt5 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase / transcription elongation / X-ray crystallography / cryo-EM / Spt4/5-DSIF-NusG | |||||||||
| Biological species |   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Bose D / Klein B / Baker K / Yusoff Z / Murakami K / Zhang X | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2011 Journal: Proc Natl Acad Sci U S A / Year: 2011Title: RNA polymerase and transcription elongation factor Spt4/5 complex structure. Authors: Brianna J Klein / Daniel Bose / Kevin J Baker / Zahirah M Yusoff / Xiaodong Zhang / Katsuhiko S Murakami /  Abstract: Spt4/5 in archaea and eukaryote and its bacterial homolog NusG is the only elongation factor conserved in all three domains of life and plays many key roles in cotranscriptional regulation and in ...Spt4/5 in archaea and eukaryote and its bacterial homolog NusG is the only elongation factor conserved in all three domains of life and plays many key roles in cotranscriptional regulation and in recruiting other factors to the elongating RNA polymerase. Here, we present the crystal structure of Spt4/5 as well as the structure of RNA polymerase-Spt4/5 complex using cryoelectron microscopy reconstruction and single particle analysis. The Spt4/5 binds in the middle of RNA polymerase claw and encloses the DNA, reminiscent of the DNA polymerase clamp and ring helicases. The transcription elongation complex model reveals that the Spt4/5 is an upstream DNA holder and contacts the nontemplate DNA in the transcription bubble. These structures reveal that the cellular RNA polymerases also use a strategy of encircling DNA to enhance its processivity as commonly observed for many nucleic acid processing enzymes including DNA polymerases and helicases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1840.map.gz emd_1840.map.gz | 257.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1840-v30.xml emd-1840-v30.xml emd-1840.xml emd-1840.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1840.tif EMD-1840.tif | 807.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1840 http://ftp.pdbj.org/pub/emdb/structures/EMD-1840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1840 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1840.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1840.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ccp4 density map for PFu RNA polymerase-Spt4-Spt5 complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pyrococcus furiosus RNA polymerase and transcription elongation f...
| Entire | Name: Pyrococcus furiosus RNA polymerase and transcription elongation factor Spt4/5 complex structure |
|---|---|
| Components |
|
-Supramolecule #1000: Pyrococcus furiosus RNA polymerase and transcription elongation f...
| Supramolecule | Name: Pyrococcus furiosus RNA polymerase and transcription elongation factor Spt4/5 complex structure type: sample / ID: 1000 Oligomeric state: Multisubunit RNAP core bound to heterodimer of Spt4 and Spt 5 Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 400 KDa / Theoretical: 400 KDa |
-Macromolecule #1: RNA polymerase core
| Macromolecule | Name: RNA polymerase core / type: protein_or_peptide / ID: 1 / Name.synonym: RNAP core / Oligomeric state: multisubunit / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Experimental: 380 KDa / Theoretical: 380 KDa |
| Recombinant expression | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
-Macromolecule #2: Spt4
| Macromolecule | Name: Spt4 / type: protein_or_peptide / ID: 2 / Name.synonym: Spt4 / Number of copies: 1 / Oligomeric state: Heterodimer with Spt5 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Experimental: 11 KDa / Theoretical: 11 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #3: Spt5
| Macromolecule | Name: Spt5 / type: protein_or_peptide / ID: 3 / Name.synonym: Spt5 / Number of copies: 1 / Oligomeric state: Heterodimer with Spt4 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Experimental: 17 KDa / Theoretical: 17 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10 mM Tris-HCl (pH 8 at 4C), 0.2M KCl, 0.1 mM EDTA, 2 mM DTT, 5 mM BME |
| Grid | Details: Glow discharged 300 mesh Quantifoil |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 90 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG/ST |
|---|---|
| Details | Low dose conditions |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS (4k x 4k) / Average electron dose: 10 e/Å2 / Details: CCD images / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Single Tilt (Gatan) / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 13.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC V, TIGRIS, EMAN / Number images used: 70890 |
| Final angle assignment | Details: IMAGIC alpha, beta and gamma |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: chimera, CNS |
| Details | Protocol: Rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name: chimera, CNS |
| Details | Protocol: Rigid body. X-ray structure of P. furiosus Spt4 and Spt5-NGN and Spt5-KOW were manually fitted into these densities. For fitting the Spt4 and Spt5-NGN, a hydrophobic cavity on the Spt5-NGN surface was positioned in front of a tip of the coiled-coil of the RpoA1 subunit as suggested by the site-directed and deletion mutagenesis studies. The model was refined using model-minimize.inp in CNS that carried out the conjugate gradient minimization with no experimental energy terms. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















