[English] 日本語
 Yorodumi
Yorodumi- EMDB-1806: Cryo-electron tomography derived density map of a conserved retro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1806 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomography derived density map of a conserved retroviral RNA packaging element from Moloney Murine Leukemia Virus. | |||||||||
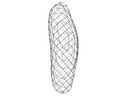
 Map data Map data | The final average of 38 subvolumes of two sets of two stem loop structures (CD2) of MoMuLV. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-ET / tomography / retroviral RNA / MoMuLV / Moloney Murine Leukemia Virus / double hairpin | |||||||||
| Biological species |  Moloney murine leukemia virus Moloney murine leukemia virus | |||||||||
| Method | subtomogram averaging / cryo EM | |||||||||
 Authors Authors | Miyazaki Y / Irobalieva RN / Tolbert B / Smalls-Mantey A / Iyalla K / Loeliger K / DSouza V / Khant H / Schmid MF / Garcia E ...Miyazaki Y / Irobalieva RN / Tolbert B / Smalls-Mantey A / Iyalla K / Loeliger K / DSouza V / Khant H / Schmid MF / Garcia E / Telesnitsky A / Chiu W / Summers MF | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2010 Journal: J Mol Biol / Year: 2010Title: Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. Authors: Yasuyuki Miyazaki / Rossitza N Irobalieva / Blanton S Tolbert / Adjoa Smalls-Mantey / Kilali Iyalla / Kelsey Loeliger / Victoria D'Souza / Htet Khant / Michael F Schmid / Eric L Garcia / ...Authors: Yasuyuki Miyazaki / Rossitza N Irobalieva / Blanton S Tolbert / Adjoa Smalls-Mantey / Kilali Iyalla / Kelsey Loeliger / Victoria D'Souza / Htet Khant / Michael F Schmid / Eric L Garcia / Alice Telesnitsky / Wah Chiu / Michael F Summers /  Abstract: The 5'-untranslated regions of all gammaretroviruses contain a conserved "double-hairpin motif" (Ψ(CD)) that is required for genome packaging. Both hairpins (SL-C and SL-D) contain GACG tetraloops ...The 5'-untranslated regions of all gammaretroviruses contain a conserved "double-hairpin motif" (Ψ(CD)) that is required for genome packaging. Both hairpins (SL-C and SL-D) contain GACG tetraloops that, in isolated RNAs, are capable of forming "kissing" interactions stabilized by two intermolecular G-C base pairs. We have determined the three-dimensional structure of the double hairpin from the Moloney murine leukemia virus ([Ψ(CD)](2), 132 nt, 42.8 kDa) using a (2)H-edited NMR-spectroscopy-based approach. This approach enabled the detection of (1)H-(1)H dipolar interactions that were not observed in previous studies of isolated SL-C and SL-D hairpin RNAs using traditional (1)H-(1)H correlated and (1)H-(13)C-edited NMR methods. The hairpins participate in intermolecular cross-kissing interactions (SL-C to SL-D' and SLC' to SL-D) and stack in an end-to-end manner (SL-C to SL-D and SL-C' to SL-D') that gives rise to an elongated overall shape (ca 95 Å×45 Å×25 Å). The global structure was confirmed by cryo-electron tomography (cryo-ET), making [Ψ(CD)](2) simultaneously the smallest RNA to be structurally characterized to date by cryo-ET and among the largest to be determined by NMR. Our findings suggest that, in addition to promoting dimerization, [Ψ(CD)](2) functions as a scaffold that helps initiate virus assembly by exposing a cluster of conserved UCUG elements for binding to the cognate nucleocapsid domains of assembling viral Gag proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1806.map.gz emd_1806.map.gz | 939.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1806-v30.xml emd-1806-v30.xml emd-1806.xml emd-1806.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1806.png 1806.png | 47.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1806 http://ftp.pdbj.org/pub/emdb/structures/EMD-1806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1806 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1806.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1806.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The final average of 38 subvolumes of two sets of two stem loop structures (CD2) of MoMuLV. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.487 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : MLV Tandem Hairpin RNA
| Entire | Name: MLV Tandem Hairpin RNA |
|---|---|
| Components |
|
-Supramolecule #1000: MLV Tandem Hairpin RNA
| Supramolecule | Name: MLV Tandem Hairpin RNA / type: sample / ID: 1000 Details: RNA synthesized by in vitro transcription and purified by polyacrylamide gel electrophoresis. Sequence of the RNA confirmed by NMR. Oligomeric state: Homodimer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 42.8 KDa / Method: Not determined but identity confirmed by NMR |
-Macromolecule #1: MLV Tandem Hairpin RNA
| Macromolecule | Name: MLV Tandem Hairpin RNA / type: rna / ID: 1 / Name.synonym: MLV Tandem Hairpin RNA / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  Moloney murine leukemia virus / synonym: Moloney Murine Leukemia Virus Moloney murine leukemia virus / synonym: Moloney Murine Leukemia Virus |
| Molecular weight | Theoretical: 42.8 KDa |
| Sequence | String: GGCGGACCCG UGGUGGAACA GACGUGUUCG GAACACCCGG CCGCAACCCU GGGAGACGUC CCAGGG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Concentration | 1.03 mg/mL |
|---|---|
| Buffer | pH: 7 Details: Tris buffer, pH 7.0, containing 140 mM KCl and 2 mM MgCl2 |
| Grid | Details: 200 mesh gold grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III / Details: Vitrification instrument: Vitrobot Mark III / Method: 1 blot, 1 second |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FSC |
|---|---|
| Temperature | Min: 98 K / Max: 98 K / Average: 98 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Specialist optics | Energy filter - Name: In column Omega-type filter |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Average electron dose: 85 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 23123 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal magnification: 20000 |
| Sample stage | Specimen holder: Gatan 70 degree holder / Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
- Image processing
Image processing
| Details | Tomogram was reconstructed using IMOD. Average number of tilts used in the 3D reconstructions: 60. Average tomographic tilt angle increment: 2. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Software - Name:  IMOD / Details: Final map was an average of 38 subvolumes. IMOD / Details: Final map was an average of 38 subvolumes. |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera, X-plor |
| Details | Protocol: Distance geometry refinement with the Cyana software package using NMR-derived restraints. The cryo-ET data were not employed as refinement restraints. The determined NMR structure (2LIF) was fitted into the cryo-ET density map using Chimera and X-plor. |
| Refinement | Space: REAL Target criteria: Lowest target functions, equals sum of the square of the distance and angle restraint violations |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















