[English] 日本語
 Yorodumi
Yorodumi- EMDB-1800: Single particle cryo-electron microscopy analysis of CD4 bound HI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1800 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryo-electron microscopy analysis of CD4 bound HIV-1 Env | |||||||||
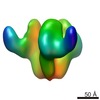
 Map data Map data | This is an image of a surface rendered CD4 bound HIV-1 Env | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 spike / Membrane Fusion / Structural Transition / Cryo-EM / Single Particle | |||||||||
| Function / homology | : / T-cell surface antigen CD4 / Human immunodeficiency virus 1, envelope glycoprotein Gp120 / viral envelope Function and homology information Function and homology information | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 21.0 Å | |||||||||
 Authors Authors | Wu SR / Loving R / Lindqvist B / Hebert H / Koeck P / Sjoberg M / Garoff H | |||||||||
 Citation Citation | Journal: AIDS Res Hum Retroviruses / Year: 1990 Title: Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. Authors: R L Garlick / R J Kirschner / F M Eckenrode / W G Tarpley / C S Tomich /  Abstract: A truncated molecule containing the N-terminal 183 amino acid residues of CD4 (sCD4-183) has been produced in Escherichia coli at high levels, using the trp promoter and an AT-rich ribosome binding ...A truncated molecule containing the N-terminal 183 amino acid residues of CD4 (sCD4-183) has been produced in Escherichia coli at high levels, using the trp promoter and an AT-rich ribosome binding site to direct expression in a pBR322-derived vector. A culture has been selected which allows large-scale fermentation and production of this material as an insoluble inclusion body protein. Procedures which solubilize, refold, and purify sCD4-183 have been developed. The purified sCD4-183 binds gp120 in solution and blocks human immunodeficiency virus (HIV) infection of human peripheral blood lymphocytes in vitro. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1800.map.gz emd_1800.map.gz | 956.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1800-v30.xml emd-1800-v30.xml emd-1800.xml emd-1800.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD_1800.jpg EMD_1800.jpg | 192.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1800 http://ftp.pdbj.org/pub/emdb/structures/EMD-1800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1800 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1800.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1800.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is an image of a surface rendered CD4 bound HIV-1 Env | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
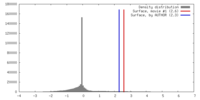
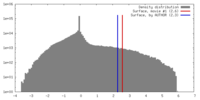
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CD4 bound HIV-1 Env
| Entire | Name: CD4 bound HIV-1 Env |
|---|---|
| Components |
|
-Supramolecule #1000: CD4 bound HIV-1 Env
| Supramolecule | Name: CD4 bound HIV-1 Env / type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: Trimer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 700 KDa / Theoretical: 500 KDa / Method: Blue Native PAGE |
-Macromolecule #1: gp160deltaCTSOS
| Macromolecule | Name: gp160deltaCTSOS / type: protein_or_peptide / ID: 1 / Name.synonym: HIV-1 Env / Number of copies: 3 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: JR-FL / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: JR-FL / synonym: HIV-1 |
| Molecular weight | Theoretical: 420 KDa |
| Recombinant expression | Organism: 293T / Recombinant plasmid: pCAGGS JRFL 160deltaCTSOS |
| Sequence | GO: viral envelope InterPro: Human immunodeficiency virus 1, envelope glycoprotein Gp120 |
-Macromolecule #2: soluble CD4 (2 domain)
| Macromolecule | Name: soluble CD4 (2 domain) / type: protein_or_peptide / ID: 2 / Name.synonym: sCD4-2d Details: sCD4-183 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 26 KDa |
| Sequence | GO: GO: 0006948 / InterPro: T-cell surface antigen CD4 |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 50 mM HEPES, 100 mM NaCl, 1.8 mM CaCl2 |
|---|---|
| Staining | Type: NEGATIVE / Details: No stain was applied |
| Grid | Details: 200 mesh Cu Holey carbon grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 77 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot / Method: Blot for 3 seconds once before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100F |
|---|---|
| Temperature | Min: 93 K / Max: 96 K / Average: 95 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected using online FFT Legacy - Electron beam tilt params: No Tilt |
| Date | Oct 23, 2009 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Digitization - Sampling interval: 3.5 µm / Number real images: 336 / Average electron dose: 9 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 43300 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | The 3D structures reconstructed by processing particle images using standard single particle procedure in EMAN version 1.9. |
|---|---|
| CTF correction | Details: Each Digitized Image |
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 21.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 9870 |
| Final two d classification | Number classes: 131 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: O |
| Details | PDBEntryID_givenInChain. Protocol: Rigid Body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)