[English] 日本語
 Yorodumi
Yorodumi- EMDB-8439: CryoEM reconstruction of hIKK1 in the most open state, conformation 1 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8439 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM reconstruction of hIKK1 in the most open state, conformation 1 | |||||||||||||||
 Map data Map data | CryoEM reconstruction of human IKK1, hexameric composition, final map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to acetate / IkappaB kinase / IkappaB kinase activity / IKBKB deficiency causes SCID / IKBKG deficiency causes anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) (via TLR) / response to cholecystokinin / SLC15A4:TASL-dependent IRF5 activation / IkappaB kinase complex / transferrin receptor binding / IkBA variant leads to EDA-ID ...response to acetate / IkappaB kinase / IkappaB kinase activity / IKBKB deficiency causes SCID / IKBKG deficiency causes anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) (via TLR) / response to cholecystokinin / SLC15A4:TASL-dependent IRF5 activation / IkappaB kinase complex / transferrin receptor binding / IkBA variant leads to EDA-ID / CD40 receptor complex / NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 / RIP-mediated NFkB activation via ZBP1 / Modulation of host responses by IFN-stimulated genes / AKT phosphorylates targets in the cytosol / response to hydroperoxide / toll-like receptor 4 signaling pathway / non-canonical NF-kappaB signal transduction / : / Constitutive Signaling by AKT1 E17K in Cancer / TRAF6 mediated NF-kB activation / positive regulation of interferon-alpha production / anatomical structure morphogenesis / canonical NF-kappaB signal transduction / response to amino acid / skeletal muscle contraction / striated muscle cell differentiation / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / TICAM1, RIP1-mediated IKK complex recruitment / MAP3K8 (TPL2)-dependent MAPK1/3 activation / tumor necrosis factor-mediated signaling pathway / IKK complex recruitment mediated by RIP1 / TNFR1-induced NF-kappa-B signaling pathway / Regulation of NF-kappa B signaling / NIK-->noncanonical NF-kB signaling / Dectin-1 mediated noncanonical NF-kB signaling / Regulation of TNFR1 signaling / Activation of NF-kappaB in B cells / : / NOD1/2 Signaling Pathway / TAK1-dependent IKK and NF-kappa-B activation / PKR-mediated signaling / cellular response to virus / response to toxic substance / CLEC7A (Dectin-1) signaling / cytoplasmic side of plasma membrane / response to virus / FCERI mediated NF-kB activation / Interleukin-1 signaling / cellular response to tumor necrosis factor / Downstream TCR signaling / ER-Phagosome pathway / scaffold protein binding / positive regulation of canonical NF-kappaB signal transduction / protein kinase activity / immune response / response to xenobiotic stimulus / inflammatory response / protein heterodimerization activity / innate immune response / protein serine/threonine kinase activity / protein-containing complex binding / SARS-CoV-2 activates/modulates innate and adaptive immune responses / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / nucleoplasm / ATP binding / cytoplasm / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
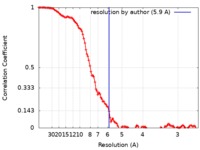
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||||||||
 Authors Authors | Lyumkis D / Ghosh G / Polley S / Biswath T / Huang D / Passos DO | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2016 Journal: Cell Rep / Year: 2016Title: Structural Basis for the Activation of IKK1/α. Authors: Smarajit Polley / Dario Oliveira Passos / De-Bin Huang / Maria Carmen Mulero / Anup Mazumder / Tapan Biswas / Inder M Verma / Dmitry Lyumkis / Gourisankar Ghosh /  Abstract: Distinct signaling pathways activate the NF-κB family of transcription factors. The canonical NF-κB-signaling pathway is mediated by IκB kinase 2/β (IKK2/β), while the non-canonical pathway ...Distinct signaling pathways activate the NF-κB family of transcription factors. The canonical NF-κB-signaling pathway is mediated by IκB kinase 2/β (IKK2/β), while the non-canonical pathway depends on IKK1/α. The structural and biochemical bases for distinct signaling by these otherwise highly similar IKKs are unclear. We report single-particle cryoelectron microscopy (cryo-EM) and X-ray crystal structures of human IKK1 in dimeric (∼150 kDa) and hexameric (∼450 kDa) forms. The hexamer, which is the representative form in the crystal but comprises only ∼2% of the particles in solution by cryo-EM, is a trimer of IKK1 dimers. While IKK1 hexamers are not detectable in cells, the surface that supports hexamer formation is critical for IKK1-dependent cellular processing of p100 to p52, the hallmark of non-canonical NF-κB signaling. Comparison of this surface to that in IKK2 indicates significant divergence, and it suggests a fundamental role for this surface in signaling by these kinases through distinct pathways. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8439.map.gz emd_8439.map.gz | 107.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8439-v30.xml emd-8439-v30.xml emd-8439.xml emd-8439.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8439_fsc.xml emd_8439_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_8439.png emd_8439.png | 121.4 KB | ||
| Others |  emd_8439_half_map_1.map.gz emd_8439_half_map_1.map.gz emd_8439_half_map_2.map.gz emd_8439_half_map_2.map.gz | 16.3 MB 16.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8439 http://ftp.pdbj.org/pub/emdb/structures/EMD-8439 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8439 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8439 | HTTPS FTP |
-Related structure data
| Related structure data |  8436C  8437C  8438C  5ebzC  5tqwC  5tqxC  5tqyC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8439.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8439.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of human IKK1, hexameric composition, final map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
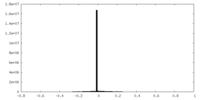
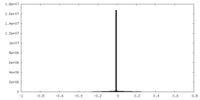
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: CryoEM reconstruction of human IKK1, hexameric composition, half...
| File | emd_8439_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of human IKK1, hexameric composition, half map 2, unfiltered and unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
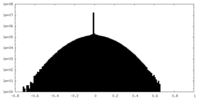
| Density Histograms |
-Half map: CryoEM reconstruction of human IKK1, hexameric composition, half...
| File | emd_8439_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of human IKK1, hexameric composition, half map 1, unfiltered and unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
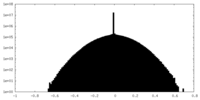
| Density Histograms |
- Sample components
Sample components
-Entire : Inhibitor of KappaB Kinase 1 hexamer
| Entire | Name: Inhibitor of KappaB Kinase 1 hexamer |
|---|---|
| Components |
|
-Supramolecule #1: Inhibitor of KappaB Kinase 1 hexamer
| Supramolecule | Name: Inhibitor of KappaB Kinase 1 hexamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: hexamer |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 450 KDa |
-Macromolecule #1: Inhibitor of Kappa B Kinase 1
| Macromolecule | Name: Inhibitor of Kappa B Kinase 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: DPEFGAGGPW EMRERLGTGG FGNVCLYQHR ELDLKIAIKS CRLELSTKNR ERWCHEIQI MKKLNHANVV KACDVPEELN ILIHDVPLLA MEYCSGGDLR K LLNKPENC CGLKESQILS LLSDIGSGIR YLHENKIIHR DLKPENIVLQ DV GGKIIHK IIDLGYAKDV ...String: DPEFGAGGPW EMRERLGTGG FGNVCLYQHR ELDLKIAIKS CRLELSTKNR ERWCHEIQI MKKLNHANVV KACDVPEELN ILIHDVPLLA MEYCSGGDLR K LLNKPENC CGLKESQILS LLSDIGSGIR YLHENKIIHR DLKPENIVLQ DV GGKIIHK IIDLGYAKDV DQGELCTEFV GTLQYLAPEL FENKPYTATV DYW SFGTMV FECIAGYRPF LHHLQPFTWH EKIKKKDPKC IFACEEMSGE VRFS SHLPQ PNSLCSLIVE PMENWLQLML NWDPQQRGGP VDLTLKQPRC FVLMD HILN LKIVHILNMT SAKIISFLLP PDESLHSLQS RIERETGINT GSQELL SET GISLDPRKPA SQCVLDGVRG CDSYMVYLFD KSKTVYEGPF ASRSLSD CV NYIVQDSKIQ LPIIQLRKVW AEAVHYVSGL KEDYSRLFQG QRAAMLSL L RYNANLTKMK NTLISASQQL KAKLEFFHKS IQLDLERYSE QMTYGISSE KMLKAWKEME EKAIHYAEVG VIGYLEDQIM SLHAEIMELQ KSPYGRRQGD LMESLEQRA IDLYKQLKHR PSDHSYSDST EMVKIIVHTV QSQDRVLKEL F GHLSKLLG CKQKIIDLLP KVEVALSNIK EADNTVMFMQ GKRQKEIWHL LK IACTQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER Details: Sample containing IKK1 dimers in SEC buffer was applied onto freshly plasma-treated (6 seconds, Gatan Solarus plasma cleaner) holey carbon C-flat grids (Protochips), adsorbed for 30 seconds, ...Details: Sample containing IKK1 dimers in SEC buffer was applied onto freshly plasma-treated (6 seconds, Gatan Solarus plasma cleaner) holey carbon C-flat grids (Protochips), adsorbed for 30 seconds, and then plunged into liquid ethane using a manual cryo-plunger in an ambient environment of 4 degrees C.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 2918 / Average exposure time: 10.0 sec. / Average electron dose: 40.0 e/Å2 Details: The dose was fractionated over 50 raw frames collected over a 10 second exposure time (200 ms per frame) on the Gatan K2 Summit direct detection device, with each frame receiving a dose of ...Details: The dose was fractionated over 50 raw frames collected over a 10 second exposure time (200 ms per frame) on the Gatan K2 Summit direct detection device, with each frame receiving a dose of ~6.5 e-/pixel/sec. 2918 movies were collected and recorded at a nominal magnification of 22,500, corresponding to a pixel size of 1.31 A at the specimen level. The individual frames were gain-corrected, then aligned and summed using a GPU-enabled whole frame alignment program (Li et al., 2013), and exposure-filtered (Grant and Grigorieff, 2015). |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 5.2 µm / Calibrated defocus min: 1.1 µm / Calibrated magnification: 38167 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Overall B value: 380 |
|---|
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)