+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1769 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
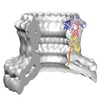
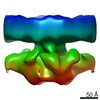
| Title | Perforin Pore | |||||||||
 Map data Map data | cryo EM reconstruction of perforin pore with 20 fold symmetry (Related to EM entries 1772 and 1773) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MACPF-CDC superfamily / pore-forming proteins | |||||||||
| Function / homology | Membrane attack complex component/perforin (MACPF) domain Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 28.5 Å | |||||||||
 Authors Authors | Lukoyanova N / Law RHP / Voskoboinik I / Caradoc-Davies TT / Baran K / Dunstone MA / D'Angelo ME / Orlova EV / Coulibaly F / Verschoor S ...Lukoyanova N / Law RHP / Voskoboinik I / Caradoc-Davies TT / Baran K / Dunstone MA / D'Angelo ME / Orlova EV / Coulibaly F / Verschoor S / Browne KA / Ciccone A / Kuiper MJ / Bird PI / Trapani JA / Whisstock JC / Saibil HR | |||||||||
 Citation Citation |  Journal: Nature / Year: 2010 Journal: Nature / Year: 2010Title: The structural basis for membrane binding and pore formation by lymphocyte perforin. Authors: Ruby H P Law / Natalya Lukoyanova / Ilia Voskoboinik / Tom T Caradoc-Davies / Katherine Baran / Michelle A Dunstone / Michael E D'Angelo / Elena V Orlova / Fasséli Coulibaly / Sandra ...Authors: Ruby H P Law / Natalya Lukoyanova / Ilia Voskoboinik / Tom T Caradoc-Davies / Katherine Baran / Michelle A Dunstone / Michael E D'Angelo / Elena V Orlova / Fasséli Coulibaly / Sandra Verschoor / Kylie A Browne / Annette Ciccone / Michael J Kuiper / Phillip I Bird / Joseph A Trapani / Helen R Saibil / James C Whisstock /  Abstract: Natural killer cells and cytotoxic T lymphocytes accomplish the critically important function of killing virus-infected and neoplastic cells. They do this by releasing the pore-forming protein ...Natural killer cells and cytotoxic T lymphocytes accomplish the critically important function of killing virus-infected and neoplastic cells. They do this by releasing the pore-forming protein perforin and granzyme proteases from cytoplasmic granules into the cleft formed between the abutting killer and target cell membranes. Perforin, a 67-kilodalton multidomain protein, oligomerizes to form pores that deliver the pro-apoptopic granzymes into the cytosol of the target cell. The importance of perforin is highlighted by the fatal consequences of congenital perforin deficiency, with more than 50 different perforin mutations linked to familial haemophagocytic lymphohistiocytosis (type 2 FHL). Here we elucidate the mechanism of perforin pore formation by determining the X-ray crystal structure of monomeric murine perforin, together with a cryo-electron microscopy reconstruction of the entire perforin pore. Perforin is a thin 'key-shaped' molecule, comprising an amino-terminal membrane attack complex perforin-like (MACPF)/cholesterol dependent cytolysin (CDC) domain followed by an epidermal growth factor (EGF) domain that, together with the extreme carboxy-terminal sequence, forms a central shelf-like structure. A C-terminal C2 domain mediates initial, Ca(2+)-dependent membrane binding. Most unexpectedly, however, electron microscopy reveals that the orientation of the perforin MACPF domain in the pore is inside-out relative to the subunit arrangement in CDCs. These data reveal remarkable flexibility in the mechanism of action of the conserved MACPF/CDC fold and provide new insights into how related immune defence molecules such as complement proteins assemble into pores. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1769.map.gz emd_1769.map.gz | 12.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1769-v30.xml emd-1769-v30.xml emd-1769.xml emd-1769.xml | 10.9 KB 10.9 KB | Display Display |  EMDB header EMDB header |
| Images |  1769image.png 1769image.png | 113.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1769 http://ftp.pdbj.org/pub/emdb/structures/EMD-1769 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1769 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1769 | HTTPS FTP |
-Validation report
| Summary document |  emd_1769_validation.pdf.gz emd_1769_validation.pdf.gz | 214.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1769_full_validation.pdf.gz emd_1769_full_validation.pdf.gz | 213.5 KB | Display | |
| Data in XML |  emd_1769_validation.xml.gz emd_1769_validation.xml.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1769 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1769 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1769 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1769 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1769.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1769.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo EM reconstruction of perforin pore with 20 fold symmetry (Related to EM entries 1772 and 1773) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Perforin pores on liposomes
| Entire | Name: Perforin pores on liposomes |
|---|---|
| Components |
|
-Supramolecule #1000: Perforin pores on liposomes
| Supramolecule | Name: Perforin pores on liposomes / type: sample / ID: 1000 Details: Pores heterogeneous in size (15-25 nm) and symmetry (18-28 subunits) Oligomeric state: 20-mer / Number unique components: 1 |
|---|
-Macromolecule #1: Perforin
| Macromolecule | Name: Perforin / type: protein_or_peptide / ID: 1 / Name.synonym: Perforin / Number of copies: 20 / Oligomeric state: 20-mer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
| Sequence | InterPro: Membrane attack complex component/perforin (MACPF) domain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.005 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 0.15M NaCl, 1mM CaCl2, 20mM Hepes |
| Grid | Details: 300, 400 lacey copper grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: OTHER / Details: Vitrification instrument: Vitrobot Method: liposomes were applied to neg. glow discharged grids first, then protein solution was added. Grids were left inside Vitrobot equilibrated at 37C for 1-2 min for pore formaition. Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 150,000x magnification |
| Date | Apr 23, 2007 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 260 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.424 µm / Nominal defocus min: 0.807 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side entry LN2 cooled, single tilt / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Image regions containing the pores with small surrounding areas of membrane were selected manually using EMAN-Boxer software |
|---|---|
| CTF correction | Details: Estimated with CTFFIND3, then phases flipped for each particle |
| Final reconstruction | Applied symmetry - Point group: C20 (20 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 28.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Imagic, Spider Details: 20-fold symmetrised reconstruction. Hand of the map has not been determined Number images used: 59 |
| Final two d classification | Number classes: 10 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  UCSF Chimera UCSF Chimera |
| Details | fitting was done manually |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















