+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Replication Protein A bound to DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Pyrococcus abyssi (archaea) / Pyrococcus abyssi (archaea) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Madru C / Martinez-Carranza M / Legrand P / Sauguet L | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: DNA-binding mechanism and evolution of replication protein A. Authors: Clément Madru / Markel Martínez-Carranza / Sébastien Laurent / Alessandra C Alberti / Maelenn Chevreuil / Bertrand Raynal / Ahmed Haouz / Rémy A Le Meur / Marc Delarue / Ghislaine ...Authors: Clément Madru / Markel Martínez-Carranza / Sébastien Laurent / Alessandra C Alberti / Maelenn Chevreuil / Bertrand Raynal / Ahmed Haouz / Rémy A Le Meur / Marc Delarue / Ghislaine Henneke / Didier Flament / Mart Krupovic / Pierre Legrand / Ludovic Sauguet /  Abstract: Replication Protein A (RPA) is a heterotrimeric single stranded DNA-binding protein with essential roles in DNA replication, recombination and repair. Little is known about the structure of RPA in ...Replication Protein A (RPA) is a heterotrimeric single stranded DNA-binding protein with essential roles in DNA replication, recombination and repair. Little is known about the structure of RPA in Archaea, the third domain of life. By using an integrative structural, biochemical and biophysical approach, we extensively characterize RPA from Pyrococcus abyssi in the presence and absence of DNA. The obtained X-ray and cryo-EM structures reveal that the trimerization core and interactions promoting RPA clustering on ssDNA are shared between archaea and eukaryotes. However, we also identified a helical domain named AROD (Acidic Rpa1 OB-binding Domain), and showed that, in Archaea, RPA forms an unanticipated tetrameric supercomplex in the absence of DNA. The four RPA molecules clustered within the tetramer could efficiently coat and protect stretches of ssDNA created by the advancing replisome. Finally, our results provide insights into the evolution of this primordial replication factor in eukaryotes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16448.map.gz emd_16448.map.gz | 327.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16448-v30.xml emd-16448-v30.xml emd-16448.xml emd-16448.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
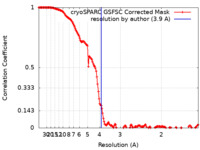
| FSC (resolution estimation) |  emd_16448_fsc.xml emd_16448_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16448.png emd_16448.png | 54.8 KB | ||
| Masks |  emd_16448_msk_1.map emd_16448_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Others |  emd_16448_half_map_1.map.gz emd_16448_half_map_1.map.gz emd_16448_half_map_2.map.gz emd_16448_half_map_2.map.gz | 322.1 MB 322.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16448 http://ftp.pdbj.org/pub/emdb/structures/EMD-16448 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16448 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16448 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16448.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16448.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.76 Å | ||||||||||||||||||||||||||||||||||||
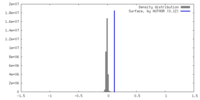
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16448_msk_1.map emd_16448_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
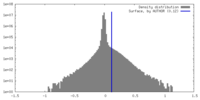
| Density Histograms |
-Half map: #1
| File | emd_16448_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
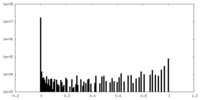
| Density Histograms |
-Half map: #2
| File | emd_16448_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
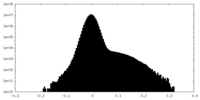
| Density Histograms |
- Sample components
Sample components
-Entire : RPA bound to a 100-mer poly-dT ssDNA
| Entire | Name: RPA bound to a 100-mer poly-dT ssDNA |
|---|---|
| Components |
|
-Supramolecule #1: RPA bound to a 100-mer poly-dT ssDNA
| Supramolecule | Name: RPA bound to a 100-mer poly-dT ssDNA / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 180 KDa |
-Supramolecule #2: RPA 1, 2 and 3
| Supramolecule | Name: RPA 1, 2 and 3 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
-Supramolecule #3: poly dT
| Supramolecule | Name: poly dT / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: RPA1
| Macromolecule | Name: RPA1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
| Recombinant expression | Organism:  |
| Sequence | String: MSVLTKDRII EIIERKTGMS REEIEEEIRK IMEEDPYLSE QGAAALLAER LGIDLIEKEE VSLMRISELY PGMDPREVNV VGRVLKKYPP REYTRKDGSV GRVASLIIYD DSGRARVVLW DAKVSEYYNK IEVGDVIKVL DAQVKESLSG LPELHINFRA RIILNPDDPR ...String: MSVLTKDRII EIIERKTGMS REEIEEEIRK IMEEDPYLSE QGAAALLAER LGIDLIEKEE VSLMRISELY PGMDPREVNV VGRVLKKYPP REYTRKDGSV GRVASLIIYD DSGRARVVLW DAKVSEYYNK IEVGDVIKVL DAQVKESLSG LPELHINFRA RIILNPDDPR VEMIPPLEEV RVATYTRKKI KDIEAGDRFV EVRGTIAKVY RVLTYDACPE CKKKVDYDEG LGVWICPEHG EVQPIKMTIL DFGLDDGTGY IRVTLFGDDA EELLGVSPEE IAEKIKELEE SGLTTKEAAR KLAEDEFYNI IGREIVVRGN VIEDRFLGLI LRASSWEDVD YRREIERIKE ELEKLGVM |
-Macromolecule #2: RPA2
| Macromolecule | Name: RPA2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
| Recombinant expression | Organism:  |
| Sequence | String: MSKKRMPATR LYIKDILEGY FVKSEGDFEP NYLITKYARK VYRAKIVGTV VREPLIAEDE TYGKFQVDDG TGVIWVLGFR DDTKFAKLVR KGDLVQVIGK IAEWRDDKQI LVEGVSKVHP NMWILHRYET LKEKIEHIKK AKIALEIYNQ YGITAKSKVI AKNKGIEEEL ...String: MSKKRMPATR LYIKDILEGY FVKSEGDFEP NYLITKYARK VYRAKIVGTV VREPLIAEDE TYGKFQVDDG TGVIWVLGFR DDTKFAKLVR KGDLVQVIGK IAEWRDDKQI LVEGVSKVHP NMWILHRYET LKEKIEHIKK AKIALEIYNQ YGITAKSKVI AKNKGIEEEL LEVIDELYGI MMEERSIEEP MEELLEEEIP EEKEENELLE KAKEDILNIL RQKRTAISRK YILKKLGDKY DEETIDDAIT ELLAQGEIYE PETGYYKLL |
-Macromolecule #3: RPA3
| Macromolecule | Name: RPA3 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
| Recombinant expression | Organism:  |
| Sequence | String: GTGDGSEVQV RRRKPAVERK ISEIREEDTR VSLIGRVIKV DKMDYMFWLD DGTGVAIIES ESDLPKVGQV VRVIGRIIRN EEGIHIYAEV IQDFSDADLE ALEEIRELER KLLPRLEGEI VW |
-Macromolecule #4: poly dT
| Macromolecule | Name: poly dT / type: dna / ID: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)